Gene
KWMTBOMO14661
Pre Gene Modal
BGIBMGA009592
Annotation
PREDICTED:_uncharacterized_protein_LOC101745081_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.339
Sequence
CDS
ATGTTTAAAATTACCGCTATCCTTTCTTTAATGCTAATCGCGACTTGGTCGCAGGCAGCGTCGCGGCGATTTAAGAAGGACGCGACAGCTTCTGATGATTTCGACGCAGGCACCACGCTCTGGGCGGCGAGTTTGTCGTGCGACTCGTGCGGTAATGAGTGCACCTCAGCATGCGGCACCAGACACTTCAGGTCGTGCTGCTTCAACTACTTGAGGAGGAAACGGGGACCCGAATCCGTCAAGATCTGGACTCCGAGTGCCCGCGACAATAACTTGGACAGCATCACTAGGCCCAGATACCCTTTCTTCGTCGTCCAGAACCTCCCCGAGCAGTGGCTGGCAAACTTGCCCTCCGATTATAAGCCTTACGATGATCTCGACACCCGACAATCGTACGACGCCTAA
Protein
MFKITAILSLMLIATWSQAASRRFKKDATASDDFDAGTTLWAASLSCDSCGNECTSACGTRHFRSCCFNYLRRKRGPESVKIWTPSARDNNLDSITRPRYPFFVVQNLPEQWLANLPSDYKPYDDLDTRQSYDA
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
BABH01036360
BABH01036361
MH028132
AXY04301.1
KQ459249
KPJ02245.1
+ More
KQ460127 KPJ17583.1 MH028131 AXY04300.1 KZ150242 PZC71905.1 KT005996 RSAL01000043 ALM30347.1 RVE50683.1 AGBW02010978 OWR47434.1 NWSH01000015 PCG80815.1 PCG80814.1 PCG80816.1 JTDY01000814 KOB75781.1 JTDY01015510 KOB51944.1 ODYU01000818 SOQ36169.1 CH477532 EAT39437.1 AXCN02002425
KQ460127 KPJ17583.1 MH028131 AXY04300.1 KZ150242 PZC71905.1 KT005996 RSAL01000043 ALM30347.1 RVE50683.1 AGBW02010978 OWR47434.1 NWSH01000015 PCG80815.1 PCG80814.1 PCG80816.1 JTDY01000814 KOB75781.1 JTDY01015510 KOB51944.1 ODYU01000818 SOQ36169.1 CH477532 EAT39437.1 AXCN02002425
Proteomes
PRIDE
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
Ontologies
Topology
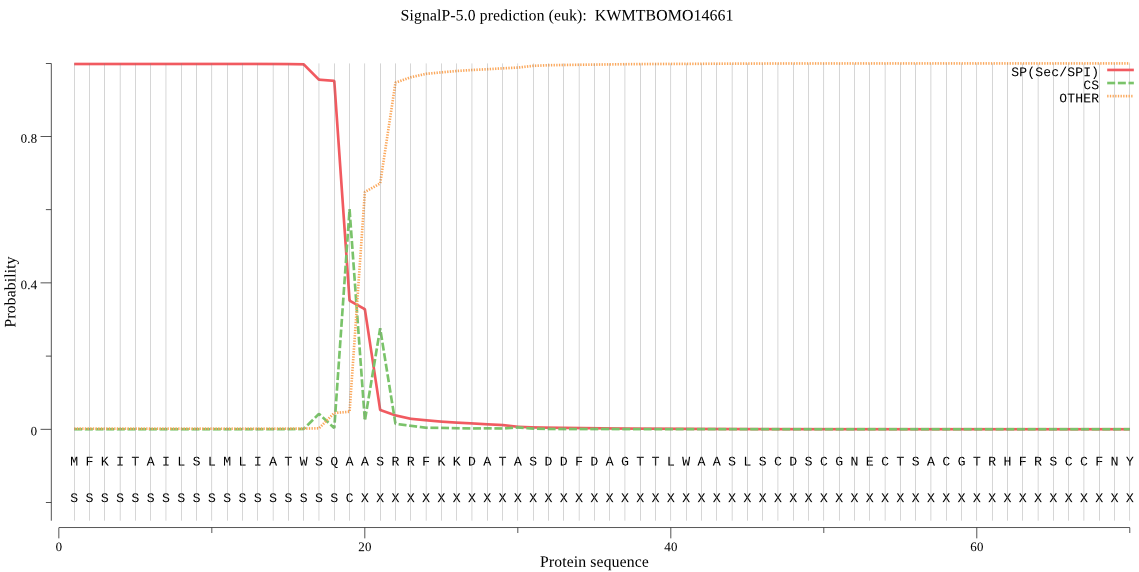
SignalP
Position: 1 - 19,
Likelihood: 0.998381
Length:
134
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.949429999999998
Exp number, first 60 AAs:
0.949279999999999
Total prob of N-in:
0.07398
outside
1 - 134

Population Genetic Test Statistics
Pi
226.114646
Theta
189.392846
Tajima's D
0.587522
CLR
0.76526
CSRT
0.536273186340683
Interpretation
Uncertain
