Gene
KWMTBOMO14654
Pre Gene Modal
BGIBMGA009595
Annotation
PREDICTED:_cleavage_stimulation_factor_subunit_1_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.777 Nuclear Reliability : 1.885
Sequence
CDS
ATGAAGGAAGACAATGTTGATATTGATACTAAAAACGTTGTTAAAAACCGCGAGCTTCTCTATCGGATGATAATAAGCCAATTGTACTATGACGGCTACCAGCCCATAGCTGCGACGTTGGCAGCAGCCGTACACGCTGACCCTCCGTGTCCTCCCAGCGACAGGTTGCTCAATGTCATGATGGTGGGCCTACAGCATGAGCCAGACAGGAAAGATAGGCTGGCTGCATCGAATGCTGCCGAGCATCTTTTGGGCACAACCGGTTTTGACTTGGAGTACGAAATGGACGCTTCCTCCCTGGCCCCTGAGCCGGCCACGTATGAGACGGCATACGTGACTTCGCACAAGATGTCTTGCCGGGCTGGAGCCTTCAGCCACTGCGGACAGCTGGTGGCCACTGGCAGTGTCGATGCCAGTATCAAGATTTTAGATGTGGAGAGGATGCTCGCGAAATCAGCTCCCGAGGAGGTGGAGCAAGGGAGGGAGCAACAGGGGCATCCCGTCATCAGAACCCTATACGACCACACTGATGAAATTACAGCGCTCGATTTTCATCCCCGAGAACAGATCTTAGTGTCGGCATCAAGGGATAACTGCATCAAGCTCTTCGACATCACCAAAGCATCAGCGAAGAAGGCCTTTAAATCAATTACGGACGCTGAACCAGTACTGTGCGTGGCCTTTCATCCCCTCGGAGATCACATCATAGCCTCGACGACACATCCGGTTATCCGGTTATATGACGTCACTACGAGTCAATGCTTCGTCTGCCCTATACCATCTCATCATCACAAGAAACACGTCAATTCTATTAAATTCTCGCCTAACGGTAAGTATTTTGCGAGCGGCAGCGCTGACGGCTCCATAAAGCTGTGGGACACCGTGTCCAATAGATGTTTCAACACGTTCACGAACGCTCACGAGGGATACGAGGTCTGCTCGGTCGCTTTCACGAAGAACAGCAAGTATCTGCTGACTTCTGGGATGGATTCCTCGATAAAACTGTGGGAACTGGCCAGCAGCAGATGCCTGATCCAATACACGGGAGCCGGAACGACCGGCAAACAGGACCACTACGCCCAGGCCGTGTTCAATCACACCGAGGACTACGTGCTCTTCCCTGACGAGGTGACGACCTCGCTCTGCACTTGGCAGTCGAGGAGCGCCAGCAGGTGCCAGCTGATGTCGCTGGGTCACAACGGGACTGTCAGGTACATAGTACATTCCGGGACCGCACCAGCTTTCCTGACCTGCAGCGACGACTACAGAGCCAGGTTTTGGTACAGGAGGAACACTCATTAG
Protein
MKEDNVDIDTKNVVKNRELLYRMIISQLYYDGYQPIAATLAAAVHADPPCPPSDRLLNVMMVGLQHEPDRKDRLAASNAAEHLLGTTGFDLEYEMDASSLAPEPATYETAYVTSHKMSCRAGAFSHCGQLVATGSVDASIKILDVERMLAKSAPEEVEQGREQQGHPVIRTLYDHTDEITALDFHPREQILVSASRDNCIKLFDITKASAKKAFKSITDAEPVLCVAFHPLGDHIIASTTHPVIRLYDVTTSQCFVCPIPSHHHKKHVNSIKFSPNGKYFASGSADGSIKLWDTVSNRCFNTFTNAHEGYEVCSVAFTKNSKYLLTSGMDSSIKLWELASSRCLIQYTGAGTTGKQDHYAQAVFNHTEDYVLFPDEVTTSLCTWQSRSASRCQLMSLGHNGTVRYIVHSGTAPAFLTCSDDYRARFWYRRNTH
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
A0A2H1VG69
A0A3S2NB76
A0A2A4K2F7
H9JJ94
A0A0N1PHC2
E2B769
+ More
A0A310SDV4 A0A195C641 A0A154PA66 A0A0L7RDG4 A0A0M8ZPS1 K7JA19 A0A2A3E3I8 A0A088ART9 E1ZYC5 A0A151XA26 A0A2J7RR69 A0A0C9RE98 A0A1Y1MU19 A0A195BD64 A0A158N8W5 F4WM24 A0A067RGL5 A0A1W4XF34 A0A026WXS7 A0A182GGX1 Q0IFW0 A0A2P8YL73 J3JVU0 A0A336KMW2 A0A336LZD0 A0A0K8TM82 A0A1Q3G101 A0A182XUY6 A0A2R7X6A9 A0A1B6M5Q2 A0A146LWW0 A0A023F5D1 A0A1B6EAQ7 A0A0V0GAI6 A0A2J7RR67 A0A182JZ10 A0A182PE04 A0A182QUL8 A0A182XIS8 Q7QCR7 A0A182HTI8 A0A182VN45 A0A182LRS0 A0A0P4VQN1 A0A182N7B8 A0A182JGE7 A0A1J1IC93 A0A1B0FHV9 A0A1A9XNA1 A0A1A9VE13 A0A1B0BS74 A0A1A9ZUN0 W8C054 A0A0A1WE28 A0A034WMG3 A0A1A9WF63 A0A182SYD5 A0A0K8U0U1 J9JK62 B4JH69 A0A0M3QXJ4 A0A2S2NSG0 B4KCH1 A0A1W4W8B2 B4M0Y3 A0A2H8TMI5 B3P517 B3MSY0 A0A2M4BP06 B4QT86 B4IJ20 A0A2M3Z954 Q9V9V0 A0A2M4AJL8 B4PP12 A0A182FFJ3 W5JLE7 A0A2M4AJR7 A0A1I8PLG9 A0A3B0JFC1 Q297R7 B4G2Y0 B4N8S7 E0VRC3 A0A1I8M6I4 C1BU05 J9JM73 A0A195FUS7 E9G5N6 L7MAR6 A0A224YQC6 A0A0N8D6H3 A0A023GJZ7 A0A0P6HF98
A0A310SDV4 A0A195C641 A0A154PA66 A0A0L7RDG4 A0A0M8ZPS1 K7JA19 A0A2A3E3I8 A0A088ART9 E1ZYC5 A0A151XA26 A0A2J7RR69 A0A0C9RE98 A0A1Y1MU19 A0A195BD64 A0A158N8W5 F4WM24 A0A067RGL5 A0A1W4XF34 A0A026WXS7 A0A182GGX1 Q0IFW0 A0A2P8YL73 J3JVU0 A0A336KMW2 A0A336LZD0 A0A0K8TM82 A0A1Q3G101 A0A182XUY6 A0A2R7X6A9 A0A1B6M5Q2 A0A146LWW0 A0A023F5D1 A0A1B6EAQ7 A0A0V0GAI6 A0A2J7RR67 A0A182JZ10 A0A182PE04 A0A182QUL8 A0A182XIS8 Q7QCR7 A0A182HTI8 A0A182VN45 A0A182LRS0 A0A0P4VQN1 A0A182N7B8 A0A182JGE7 A0A1J1IC93 A0A1B0FHV9 A0A1A9XNA1 A0A1A9VE13 A0A1B0BS74 A0A1A9ZUN0 W8C054 A0A0A1WE28 A0A034WMG3 A0A1A9WF63 A0A182SYD5 A0A0K8U0U1 J9JK62 B4JH69 A0A0M3QXJ4 A0A2S2NSG0 B4KCH1 A0A1W4W8B2 B4M0Y3 A0A2H8TMI5 B3P517 B3MSY0 A0A2M4BP06 B4QT86 B4IJ20 A0A2M3Z954 Q9V9V0 A0A2M4AJL8 B4PP12 A0A182FFJ3 W5JLE7 A0A2M4AJR7 A0A1I8PLG9 A0A3B0JFC1 Q297R7 B4G2Y0 B4N8S7 E0VRC3 A0A1I8M6I4 C1BU05 J9JM73 A0A195FUS7 E9G5N6 L7MAR6 A0A224YQC6 A0A0N8D6H3 A0A023GJZ7 A0A0P6HF98
Pubmed
19121390
26354079
20798317
20075255
28004739
21347285
+ More
21719571 24845553 24508170 30249741 26483478 17510324 29403074 22516182 23537049 26369729 25244985 26823975 25474469 12364791 14747013 17210077 27129103 24495485 25830018 25348373 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20920257 23761445 15632085 20566863 25315136 21292972 25576852 28797301
21719571 24845553 24508170 30249741 26483478 17510324 29403074 22516182 23537049 26369729 25244985 26823975 25474469 12364791 14747013 17210077 27129103 24495485 25830018 25348373 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20920257 23761445 15632085 20566863 25315136 21292972 25576852 28797301
EMBL
ODYU01002410
SOQ39853.1
RSAL01000128
RVE46489.1
NWSH01000206
PCG78427.1
+ More
BABH01036343 KQ460857 KPJ11822.1 GL446149 EFN88430.1 KQ760869 OAD58752.1 KQ978220 KYM96317.1 KQ434856 KZC08717.1 KQ414614 KOC68844.1 KQ438551 KOX67153.1 AAZX01001957 AAZX01016730 KZ288427 PBC25862.1 GL435204 EFN73824.1 KQ982353 KYQ57189.1 NEVH01000611 PNF43331.1 GBYB01014919 JAG84686.1 GEZM01021115 JAV89131.1 KQ976519 KYM82139.1 ADTU01001363 GL888217 EGI64668.1 KK852521 KDR22133.1 KK107064 QOIP01000008 EZA60842.1 RLU19496.1 JXUM01062620 KQ562208 KXJ76407.1 CH477286 EAT44740.1 PYGN01000515 PSN45009.1 APGK01037334 BT127358 KB740948 KB632288 AEE62320.1 ENN77307.1 ERL91547.1 UFQS01000646 UFQT01000646 SSX05707.1 SSX26066.1 UFQS01000316 UFQT01000316 SSX02799.1 SSX23170.1 GDAI01002362 JAI15241.1 GFDL01001564 JAV33481.1 KK857471 PTY27201.1 GEBQ01008727 JAT31250.1 GDHC01007482 JAQ11147.1 GBBI01002384 JAC16328.1 GEDC01002318 JAS34980.1 GECL01001171 JAP04953.1 PNF43330.1 AXCN02000146 AAAB01008859 EAA07802.3 APCN01000536 AXCM01004364 GDKW01001814 JAI54781.1 CVRI01000047 CRK97819.1 CCAG010009819 JXJN01019545 GAMC01001518 JAC05038.1 GBXI01017361 JAC96930.1 GAKP01003592 JAC55360.1 GDHF01032161 JAI20153.1 ABLF02009770 CH916369 EDV92760.1 CP012526 ALC45969.1 GGMR01007490 MBY20109.1 CH933806 EDW14790.1 CH940650 EDW67394.1 GFXV01003067 MBW14872.1 CH954182 EDV52931.1 CH902623 EDV30370.1 GGFJ01005601 MBW54742.1 CM000364 EDX15148.1 CH480846 EDW50973.1 GGFM01004281 MBW25032.1 AE014297 AY094802 AAF57183.1 AAM11155.1 GGFK01007659 MBW40980.1 CM000160 EDW99248.1 ADMH02001223 ETN63599.1 GGFK01007708 MBW41029.1 OUUW01000005 SPP81027.1 CM000070 EAL28138.1 CH479179 EDW24175.1 CH964232 EDW81528.1 DS235465 EEB15929.1 BT078084 HACA01005879 ACO12508.1 CDW23240.1 ABLF02024491 KQ981264 KYN44042.1 GL732532 EFX85597.1 GACK01004776 JAA60258.1 GFPF01006813 MAA17959.1 GDIP01064063 LRGB01000568 JAM39652.1 KZS17962.1 GBBM01001156 JAC34262.1 GDIQ01020270 JAN74467.1
BABH01036343 KQ460857 KPJ11822.1 GL446149 EFN88430.1 KQ760869 OAD58752.1 KQ978220 KYM96317.1 KQ434856 KZC08717.1 KQ414614 KOC68844.1 KQ438551 KOX67153.1 AAZX01001957 AAZX01016730 KZ288427 PBC25862.1 GL435204 EFN73824.1 KQ982353 KYQ57189.1 NEVH01000611 PNF43331.1 GBYB01014919 JAG84686.1 GEZM01021115 JAV89131.1 KQ976519 KYM82139.1 ADTU01001363 GL888217 EGI64668.1 KK852521 KDR22133.1 KK107064 QOIP01000008 EZA60842.1 RLU19496.1 JXUM01062620 KQ562208 KXJ76407.1 CH477286 EAT44740.1 PYGN01000515 PSN45009.1 APGK01037334 BT127358 KB740948 KB632288 AEE62320.1 ENN77307.1 ERL91547.1 UFQS01000646 UFQT01000646 SSX05707.1 SSX26066.1 UFQS01000316 UFQT01000316 SSX02799.1 SSX23170.1 GDAI01002362 JAI15241.1 GFDL01001564 JAV33481.1 KK857471 PTY27201.1 GEBQ01008727 JAT31250.1 GDHC01007482 JAQ11147.1 GBBI01002384 JAC16328.1 GEDC01002318 JAS34980.1 GECL01001171 JAP04953.1 PNF43330.1 AXCN02000146 AAAB01008859 EAA07802.3 APCN01000536 AXCM01004364 GDKW01001814 JAI54781.1 CVRI01000047 CRK97819.1 CCAG010009819 JXJN01019545 GAMC01001518 JAC05038.1 GBXI01017361 JAC96930.1 GAKP01003592 JAC55360.1 GDHF01032161 JAI20153.1 ABLF02009770 CH916369 EDV92760.1 CP012526 ALC45969.1 GGMR01007490 MBY20109.1 CH933806 EDW14790.1 CH940650 EDW67394.1 GFXV01003067 MBW14872.1 CH954182 EDV52931.1 CH902623 EDV30370.1 GGFJ01005601 MBW54742.1 CM000364 EDX15148.1 CH480846 EDW50973.1 GGFM01004281 MBW25032.1 AE014297 AY094802 AAF57183.1 AAM11155.1 GGFK01007659 MBW40980.1 CM000160 EDW99248.1 ADMH02001223 ETN63599.1 GGFK01007708 MBW41029.1 OUUW01000005 SPP81027.1 CM000070 EAL28138.1 CH479179 EDW24175.1 CH964232 EDW81528.1 DS235465 EEB15929.1 BT078084 HACA01005879 ACO12508.1 CDW23240.1 ABLF02024491 KQ981264 KYN44042.1 GL732532 EFX85597.1 GACK01004776 JAA60258.1 GFPF01006813 MAA17959.1 GDIP01064063 LRGB01000568 JAM39652.1 KZS17962.1 GBBM01001156 JAC34262.1 GDIQ01020270 JAN74467.1
Proteomes
UP000283053
UP000218220
UP000005204
UP000053240
UP000008237
UP000078542
+ More
UP000076502 UP000053825 UP000053105 UP000002358 UP000242457 UP000005203 UP000000311 UP000075809 UP000235965 UP000078540 UP000005205 UP000007755 UP000027135 UP000192223 UP000053097 UP000279307 UP000069940 UP000249989 UP000008820 UP000245037 UP000019118 UP000030742 UP000076408 UP000075881 UP000075885 UP000075886 UP000076407 UP000007062 UP000075840 UP000075903 UP000075883 UP000075884 UP000075880 UP000183832 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000075901 UP000007819 UP000001070 UP000092553 UP000009192 UP000192221 UP000008792 UP000008711 UP000007801 UP000000304 UP000001292 UP000000803 UP000002282 UP000069272 UP000000673 UP000095300 UP000268350 UP000001819 UP000008744 UP000007798 UP000009046 UP000095301 UP000078541 UP000000305 UP000076858
UP000076502 UP000053825 UP000053105 UP000002358 UP000242457 UP000005203 UP000000311 UP000075809 UP000235965 UP000078540 UP000005205 UP000007755 UP000027135 UP000192223 UP000053097 UP000279307 UP000069940 UP000249989 UP000008820 UP000245037 UP000019118 UP000030742 UP000076408 UP000075881 UP000075885 UP000075886 UP000076407 UP000007062 UP000075840 UP000075903 UP000075883 UP000075884 UP000075880 UP000183832 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000075901 UP000007819 UP000001070 UP000092553 UP000009192 UP000192221 UP000008792 UP000008711 UP000007801 UP000000304 UP000001292 UP000000803 UP000002282 UP000069272 UP000000673 UP000095300 UP000268350 UP000001819 UP000008744 UP000007798 UP000009046 UP000095301 UP000078541 UP000000305 UP000076858
Pfam
Interpro
IPR017986
WD40_repeat_dom
+ More
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR038184 CSTF1_dimer_sf
IPR019775 WD40_repeat_CS
IPR032028 CSTF1_dimer
IPR036322 WD40_repeat_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR001254 Trypsin_dom
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR031945 DUF4775
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR011047 Quinoprotein_ADH-like_supfam
IPR000719 Prot_kinase_dom
IPR024977 Apc4_WD40_dom
IPR008010 Tatp1
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR038184 CSTF1_dimer_sf
IPR019775 WD40_repeat_CS
IPR032028 CSTF1_dimer
IPR036322 WD40_repeat_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR001254 Trypsin_dom
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR031945 DUF4775
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR011047 Quinoprotein_ADH-like_supfam
IPR000719 Prot_kinase_dom
IPR024977 Apc4_WD40_dom
IPR008010 Tatp1
Gene 3D
CDD
ProteinModelPortal
A0A2H1VG69
A0A3S2NB76
A0A2A4K2F7
H9JJ94
A0A0N1PHC2
E2B769
+ More
A0A310SDV4 A0A195C641 A0A154PA66 A0A0L7RDG4 A0A0M8ZPS1 K7JA19 A0A2A3E3I8 A0A088ART9 E1ZYC5 A0A151XA26 A0A2J7RR69 A0A0C9RE98 A0A1Y1MU19 A0A195BD64 A0A158N8W5 F4WM24 A0A067RGL5 A0A1W4XF34 A0A026WXS7 A0A182GGX1 Q0IFW0 A0A2P8YL73 J3JVU0 A0A336KMW2 A0A336LZD0 A0A0K8TM82 A0A1Q3G101 A0A182XUY6 A0A2R7X6A9 A0A1B6M5Q2 A0A146LWW0 A0A023F5D1 A0A1B6EAQ7 A0A0V0GAI6 A0A2J7RR67 A0A182JZ10 A0A182PE04 A0A182QUL8 A0A182XIS8 Q7QCR7 A0A182HTI8 A0A182VN45 A0A182LRS0 A0A0P4VQN1 A0A182N7B8 A0A182JGE7 A0A1J1IC93 A0A1B0FHV9 A0A1A9XNA1 A0A1A9VE13 A0A1B0BS74 A0A1A9ZUN0 W8C054 A0A0A1WE28 A0A034WMG3 A0A1A9WF63 A0A182SYD5 A0A0K8U0U1 J9JK62 B4JH69 A0A0M3QXJ4 A0A2S2NSG0 B4KCH1 A0A1W4W8B2 B4M0Y3 A0A2H8TMI5 B3P517 B3MSY0 A0A2M4BP06 B4QT86 B4IJ20 A0A2M3Z954 Q9V9V0 A0A2M4AJL8 B4PP12 A0A182FFJ3 W5JLE7 A0A2M4AJR7 A0A1I8PLG9 A0A3B0JFC1 Q297R7 B4G2Y0 B4N8S7 E0VRC3 A0A1I8M6I4 C1BU05 J9JM73 A0A195FUS7 E9G5N6 L7MAR6 A0A224YQC6 A0A0N8D6H3 A0A023GJZ7 A0A0P6HF98
A0A310SDV4 A0A195C641 A0A154PA66 A0A0L7RDG4 A0A0M8ZPS1 K7JA19 A0A2A3E3I8 A0A088ART9 E1ZYC5 A0A151XA26 A0A2J7RR69 A0A0C9RE98 A0A1Y1MU19 A0A195BD64 A0A158N8W5 F4WM24 A0A067RGL5 A0A1W4XF34 A0A026WXS7 A0A182GGX1 Q0IFW0 A0A2P8YL73 J3JVU0 A0A336KMW2 A0A336LZD0 A0A0K8TM82 A0A1Q3G101 A0A182XUY6 A0A2R7X6A9 A0A1B6M5Q2 A0A146LWW0 A0A023F5D1 A0A1B6EAQ7 A0A0V0GAI6 A0A2J7RR67 A0A182JZ10 A0A182PE04 A0A182QUL8 A0A182XIS8 Q7QCR7 A0A182HTI8 A0A182VN45 A0A182LRS0 A0A0P4VQN1 A0A182N7B8 A0A182JGE7 A0A1J1IC93 A0A1B0FHV9 A0A1A9XNA1 A0A1A9VE13 A0A1B0BS74 A0A1A9ZUN0 W8C054 A0A0A1WE28 A0A034WMG3 A0A1A9WF63 A0A182SYD5 A0A0K8U0U1 J9JK62 B4JH69 A0A0M3QXJ4 A0A2S2NSG0 B4KCH1 A0A1W4W8B2 B4M0Y3 A0A2H8TMI5 B3P517 B3MSY0 A0A2M4BP06 B4QT86 B4IJ20 A0A2M3Z954 Q9V9V0 A0A2M4AJL8 B4PP12 A0A182FFJ3 W5JLE7 A0A2M4AJR7 A0A1I8PLG9 A0A3B0JFC1 Q297R7 B4G2Y0 B4N8S7 E0VRC3 A0A1I8M6I4 C1BU05 J9JM73 A0A195FUS7 E9G5N6 L7MAR6 A0A224YQC6 A0A0N8D6H3 A0A023GJZ7 A0A0P6HF98
PDB
6B3X
E-value=1.33793e-118,
Score=1091
Ontologies
GO
PANTHER
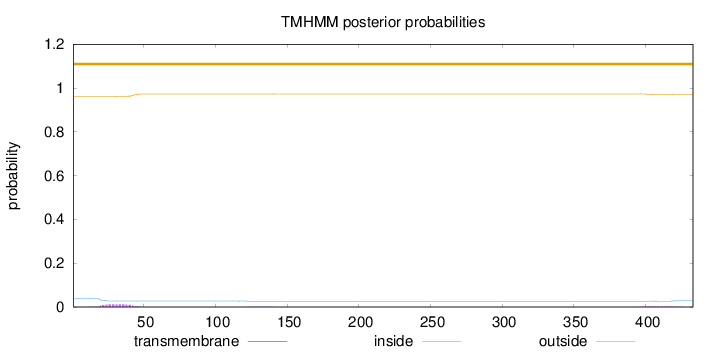
Topology
Length:
433
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.31694
Exp number, first 60 AAs:
0.26729
Total prob of N-in:
0.03923
outside
1 - 433
Population Genetic Test Statistics
Pi
274.93267
Theta
166.892645
Tajima's D
1.92476
CLR
0.137313
CSRT
0.871456427178641
Interpretation
Uncertain
