Gene
KWMTBOMO14653
Pre Gene Modal
BGIBMGA003354
Annotation
hypothetical_protein_ANCDUO_02957_[Ancylostoma_duodenale]
Full name
ATP-dependent DNA helicase
Location in the cell
Nuclear Reliability : 2.634
Sequence
CDS
ATGCCACGCAAACGTAAATCCAATCTAAGCCGTAGTTCTAGCAGAGCTCGAACTGCAAAAGTTGCTAGAAGCCAAGAATCTGAAGAGCAGACTCAGACGCGTCAGATGTTAGATTCTCAGCGTCATGCGACTCAAAGAGCTTCTGAGACCTATACGCAGACTCAAGCCCGTAGGACCTTAGATGCTGAGCGCCACGCATCTCAAAGGGCCTCTGAGACGTTTACACAGACTCAAGCCCGTCAGGCCCTAGATGCTGAACGCCACGCATCTCAAAGAGCCTCTGAGACGTTTACGCAGACTGAGACCCGTCAGGCCCTGGATGCTGAACGCCACGCATCTCAAAGGGCCTCTGAGACCTATACGCAGACTCAAGCCCGACAAACCTTAGATGCTGAGCGCCACGCATTTCAAAGGGCCTCTGAGACGTTTACGCAGACTCAGGCCCGTCAGACCCTGGATGCTGAACGCCATGCATCTCAAAGAGCTGCTGAGACAGTGGAACAGACTCAGACACGTCGAGTTTTAGATGCCGAACGTCACGCGCAATTGATTGCGGCAGAGACTGACGAAGATTCACAAAGACGACGTCTTTTAAATGCTGAACGGCAGGCAGAAAGAAGACGTACGTTTTCAATGAGTACGTGGGAGGCATTTAATGGTGCCGCTTTTGAGTATGACCCTTTGATCGACTATGTTAATCACCGATTGGTTATCATTGGGAGAATGGATAAAAAATGCAGATATTGTGAAGCACTGAAATGGAAAGAAGAAACTCCAGCACTAGAAAGAATGCCGGGAGAGGATTATAGGTTGGTGATGCACCCCGATCGCACACCAAGTGGGGAACATGAAAGGCGGTATAATGCACCGTTAATAAATGAAGTCGCAGCCTTGGTTACAGGCGAACAGTTTGCTTTCCGTGATATAGTTTTACAGGCCCGAGATGACACGTTAACCAGGGTGCCTGATACTCATAAATTTTATGATGCGTTGCAATATCCGATCATATTTAGTAAGGGACAAGAGGGGTACCACTTTCAAGTTCCTCAAATTAATCCTGTAACAGGTCTTCCCTTGCATAACAAAAAGGTTTCATGCATGGATTTTTATGCCTACAACATGATGATACGAGAAAACAACTTTAACATTGTACAAAGATGCAAGCAACTGGCCAATCAGTTTTACGTTGACATGTACGTTAAAGTTGAAAGCGAGAGGTTACGGTACATATCATTAAATCAAACTAAACTAAGAGCAGAGAATTACATCCATCTTCAAGACGCTGTGGCAAATGACGCTAATTTAAATCCAAATAACTTAGGTCGTATGGTCATTTTACCATCTTCATTTGTAAATAGCCCGCGGTACTTACACGAATATACTCAGGACGCGTTTGTTTATGTGCGTACACATGATCGCCCTGACTTATTCGTCACCTTTACTTGCAACCCAGCCTGGCCCGAAATAGTCAATCAGTTGATGCCGGGGCAAAGCGCAATAGATAGAGATGATGTAGTCGCAAGAGTTTTTAGACTAAAAGTTAAAAAACTAATGGATGTGATTAATAAAGGCCGAGTGTTCAGAGAAGCGCGCTGCTTTATGTACTCCGTCGAATGGCAGAAACGTGGACTGCCACATGTCCATATATTATTGTGGTTAAAAGATAAGTTACGACCCGATCAAATAGATAACATAATCAGCGCTGAAATACCGGATCCTAACATTGACAAGACTCTCCATGACGTTATTGTAAAAAATATGATTCACGGTCCATGCGGACCCGAAAATCCACAGTGCCACTGCATGAAAGACGGAAAATGCACAAAAAAATTTCCTCGCAAATTACAAGAGCAAAATCCCAATGTCACGTACAGTGATTTAATTTACAATGAGGGTCTGACAAAAATTGAAGACCAGGTTAAAACAATTTCTAGAAAGGATTTATCAGATTATGGAATGATCAGACCACAGAGAACCGAGGAAATCAGTAGCGATTTAATACGGGAACTAGATTACGATACTGCTTCCTTACAACAACAATTGACAGAGTCTGTTCCACGTTTGATCCCAGAACAAAGGTTAGTATTTGACAATGTTTTCAAAAAAATCGAAGATGGCGAAGGTGGTTTGTTATTTTTGGATGCTCCCGGAGGCACGGGAAAAACCTTTCTTTTAAACTTGCTTTTAGCGCAAATCCGAAAAGATAAAGGAATTGCTGTAGCAGTTGCCTCTTTCGGAATAGCCGCCACTCTGCTCAATGGTGGTCGAACGGCACATTCGGTGCTTAAACTGCCATTAAACTTAGCCCATGAAGAAATGCCAGTCTGTAATATAAGTAAAAACAGCGAGCGTGGACGTATGTTGCAGCAATGTAAATTATTAGTTTGGGATGAGTGTACTATGTCCCATAAAAGAGCAATTGAAGCTCTAGATCGGACTTTGAAAGACATCAAGAGTAATCCAAATATCATGGGGGGGATGGTGGTACTTTTAGCGGGCGATTTTAGACAGACTTTGCCGGTTATCACTAAAGGCACCCCTGCAGATGAAATAAATGCGTGTTTAAAAGCGTCAACGTTATGGGTGCACGTAAAAACGTTTAGTCTGTCTACAAATATGAGAGTGCAACTACACAATGATGTACAATCTGGACAATATGCTGCTGCTCTTCTTAAAATTGGAGAAGATCGTATGGCAAAGGATGGTAATGGCATGATTACATTGGATCGTGAATTCTGCAATGTTGTGCATAATCCAGATGATCTAAATAACTTCGTCTATAGCGAACTGCTAACTAATATGAGGAATAGAGACTGGTTATGTGAAAGAGCAATTTTAGGGCCGACGAATGAAATGGTGGGACAGATAAACGAGCAAATTATGTCACGTGTGGAAGGTGATATTGTTGAGTTTCTCTCTGTAGACACTGTAATGGATACTGAACAAGTAACATCATACCCTGTAGAATTTCTTAATTCTTTAGAACTGTTGGGAGTACCTTCACACAAACTGAGGCTGAAAGTAGGCGTTACTGTCCTTTTAATTCGGAATTTAGATGCACCAAGACTGTGTAATGGCACACGGTTGCAAGTGACACACCTCGGTCGCAATATTGTAAAAGCAATCATTATGACTGGAATGGCAAAAGGAGAGAACGTTTTAATCCCCCGTATACCCATAATTCCGACAGATTTGCCATTCCAGTTTAAGAGATTACAGTTTCCGCTGAAAATCGCATTCGCTATGACGATAAATAAAGCTCAGGGGCAAACATTAAAAGTAGCCGGCATTAATTTAGAAAAGAGTTGTTTTTCTCACGGACAATTATACGTGGCTTGCTCACGTGTTTCAAGTCCACAGAATCTCCATGTGTTGGCCAGAGAGGGAAAAACAAAGAACATAGTTTACAAAAATGTACTACAGTAA
Protein
MPRKRKSNLSRSSSRARTAKVARSQESEEQTQTRQMLDSQRHATQRASETYTQTQARRTLDAERHASQRASETFTQTQARQALDAERHASQRASETFTQTETRQALDAERHASQRASETYTQTQARQTLDAERHAFQRASETFTQTQARQTLDAERHASQRAAETVEQTQTRRVLDAERHAQLIAAETDEDSQRRRLLNAERQAERRRTFSMSTWEAFNGAAFEYDPLIDYVNHRLVIIGRMDKKCRYCEALKWKEETPALERMPGEDYRLVMHPDRTPSGEHERRYNAPLINEVAALVTGEQFAFRDIVLQARDDTLTRVPDTHKFYDALQYPIIFSKGQEGYHFQVPQINPVTGLPLHNKKVSCMDFYAYNMMIRENNFNIVQRCKQLANQFYVDMYVKVESERLRYISLNQTKLRAENYIHLQDAVANDANLNPNNLGRMVILPSSFVNSPRYLHEYTQDAFVYVRTHDRPDLFVTFTCNPAWPEIVNQLMPGQSAIDRDDVVARVFRLKVKKLMDVINKGRVFREARCFMYSVEWQKRGLPHVHILLWLKDKLRPDQIDNIISAEIPDPNIDKTLHDVIVKNMIHGPCGPENPQCHCMKDGKCTKKFPRKLQEQNPNVTYSDLIYNEGLTKIEDQVKTISRKDLSDYGMIRPQRTEEISSDLIRELDYDTASLQQQLTESVPRLIPEQRLVFDNVFKKIEDGEGGLLFLDAPGGTGKTFLLNLLLAQIRKDKGIAVAVASFGIAATLLNGGRTAHSVLKLPLNLAHEEMPVCNISKNSERGRMLQQCKLLVWDECTMSHKRAIEALDRTLKDIKSNPNIMGGMVVLLAGDFRQTLPVITKGTPADEINACLKASTLWVHVKTFSLSTNMRVQLHNDVQSGQYAAALLKIGEDRMAKDGNGMITLDREFCNVVHNPDDLNNFVYSELLTNMRNRDWLCERAILGPTNEMVGQINEQIMSRVEGDIVEFLSVDTVMDTEQVTSYPVEFLNSLELLGVPSHKLRLKVGVTVLLIRNLDAPRLCNGTRLQVTHLGRNIVKAIIMTGMAKGENVLIPRIPIIPTDLPFQFKRLQFPLKIAFAMTINKAQGQTLKVAGINLEKSCFSHGQLYVACSRVSSPQNLHVLAREGKTKNIVYKNVLQ
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
EC Number
3.6.4.12
EMBL
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
PDB
5FHH
E-value=8.87618e-20,
Score=243
Ontologies
GO
Topology
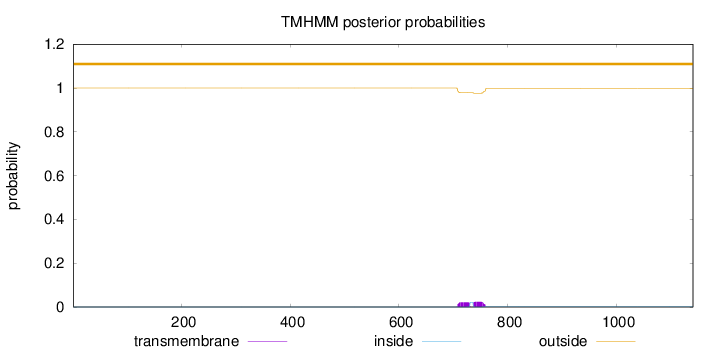
Length:
1141
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.95458
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00029
outside
1 - 1141
Population Genetic Test Statistics
Pi
352.228829
Theta
222.457772
Tajima's D
3.661507
CLR
0.159389
CSRT
0.995700214989251
Interpretation
Possibly Balancing Selection
