Gene
KWMTBOMO14651
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.988 Nuclear Reliability : 1.716
Sequence
CDS
ATGATTGCACACCGCAGAACCAGGCCCCTAGTCCCCGGCGGCCCCGCTATTCCTGGTTACGGTAGCGGCCATGGTAACGGCGGGGCAAGGGGTGCTAAGAATCCCCGGCAGAGATCCGGCTACCACCCCCCTTCACAACGACAACTGGCCCTGGTGACATACAACGTGCGCACTTTGAGGTTGGACGAGAAGCTAATAGAGCTGGAAGAAGAAATGAACAAGTTGCGCTGGGACGTTATTGGTTTATCGGAGATCCGAAGAGAGGGGGAGGATACGAAAACCTTACAGTCCGGCCACTTGTTCTACTACCGGGAGGGAGAACAGAAGTCCCAAGGTGGTGTTGGGTTTATCGTCCACAAATCTCTCATAAACAACATTGTGGCCATCGAAAGTGTGTCGAGTAGGGTAGTATACCTTGTCCTCAAAATCTCAAAGCGGTATTCGCTGAAGATTGTACAGGTATACGCCCCATCAACGTCACATCCAGGCGATGAAGTGGCAGCTACGTATGAGGACATTAGTAGGGCCATACGTAACTCCCAATCACATTTTACCATTGTTATGGGAGATTTCAATGCAAAACTGGGCCGTAGAGGCGATGATGAGTTGAAAGTGGGGCCATTTGGATTTGGGCAGCGGAATCCCAGAGGACAGATGCTGGCGGACTTTATGGAGAAGGAAGGACTCTTTATGATGAATTCTTTCTTCAAGAAGCCGCCACAACGGAAATGGACCTGGTTGAGTCCCGACGGTGTAACAAGAAATGAAATCGATTTCATCATGAGCACCAACAGACAGATATTCAACGATGTCTCTGTGATCAACAGTGTTAAAACAGGAAGTGATCACCGAATCGTCAGAGGCATGTTGAATATCAATGTCAGACTGGAAAGATCGCGTCTGATGAAGTCAACGCTTCGACCGTCGAATGCACAAATCCAAAACCCCGAGAGCTTTCAACTCGAATTGGCTAATCGCTTCGAATGGCTAAACAGCTGCGCATCAGTGGACGATGTAAACAGCAGGCTGAGACGCTTGCAAAGCCAGCTGACGAAGCTGAAGACCGAAGATGGACGTGTAACGTCGAATAAGTCAGAGGTCCTGCGGGAGATAGAAAGGTTCTACGGGCAGCTGTTTACCTCGGTCTCAGCTGAACCGCAAGGTTTAGCTGCAGACCCTAGAGCCAAGCTAACCCGACACTACACCGAAGACATCCCGGACATCAGCCTGTACGAGATTAGGATGGCTCTCAAACAGCTTAAGAATAACAAGGCACCGGGCGAGGATGGAATTACTACGGAGCTTCTGAAAGCGGGCGGGACGCCGGTACTGAAAGTCCTTCAGAAGCTCTTCAATTCCTCTTTGCTCGATGGAAAACCGCCACAGGCATGGAGCAGGGGTGTTGTGATCCTGTTCTTCAAAAAAGGTGACAAAACCTCATTGAAGAACTATAGGCCCATAGCACTGCTGAGCCATATCTACAAGCTGTTCTCGAGAGTCATCACGAAGCGTCTCGAACGCAGGTTTGATGACTTCCAGCCTACCGAACAAGCCGGTTTCCGAAAAGGCTACAGTACCATAGACCACATACATACGCTGCGGCAGATTGTACAGAAGACCGAAGAGTACAATCGGCCATTATGCTTAGCGTTTGTGGACTATGAGAAAGCCTTTGATTCCGTCGAAACATGGGCGGTCCTTAGATCTCTCCAGCGGTGCCGTATTGACCACCGGTACATCGAAGTATTGAAGTGTTTGTATAATAACGCCACCATGTCAGTCCGAGTACAGGAGCATTGCACGAAGGAAATCCCTGTAAAGAGAGGAGTGAGACAGGGAGATGTGATATCTCCGAAACTGTTTATCACTGCTCTGGAGGATTCCTTCAAGCTTCTGGAATGGCAAGGACTTGGCATCAATATTAACGACGAATACATCACTCATCTTCGGTTTGCCGATGACATCGTGGTCATGGCAGAGTCGCTGGAAGATTTAGGGCGTTTGCTCGGTGACCTCAGTAGGGTTTCTCAACAAGTGGGTCTTAAAATGAACATGGACAAAACAAAAATCATGTACAATGTCCATGTTGCACCCACCATAGTTACTGTTGGGAGCTCTACTCTTGAAGTTGTTGACGAATATATATACCTAGGACAGGCAGTCCGATTAGGTAGGTCCAACTTTGAGAGAGAGGTCAGTAGACGGATCCAACTCGGCTGGGCAGCGTTCGGTAAACTGCACAACGTCTTCTCGTCCAAAATACCTCAGTGTCTTAAGACAAAGGCGTACAACCAATGTGTGTTACCAGTGATGACTTACGGCTCGGAGACGTGGTGCTTCACTAGGGATCTTTTTAGAAGGCTCAAAGTCGCGCAGCGAGCTATGGAGAGGGCTATGCTCGGCATTTCTCTACGTGATCGAATCCGAAATGAGAAGATCCGTAGAAGAACGAAGGTTGCTGACATAGCCCGTAAAATTAGTGAACTGAAGTGGCAATGGGCCGGCCACATAGCACGAAGAGAGGACGGTCGATGGGGCAGAAAGATCCTGGAGTGGCGACCACGTACTGGCAAGCGCAGTGTGGACGGCCACCTACTAGATGGACCGACGACCTAG
Protein
MIAHRRTRPLVPGGPAIPGYGSGHGNGGARGAKNPRQRSGYHPPSQRQLALVTYNVRTLRLDEKLIELEEEMNKLRWDVIGLSEIRREGEDTKTLQSGHLFYYREGEQKSQGGVGFIVHKSLINNIVAIESVSSRVVYLVLKISKRYSLKIVQVYAPSTSHPGDEVAATYEDISRAIRNSQSHFTIVMGDFNAKLGRRGDDELKVGPFGFGQRNPRGQMLADFMEKEGLFMMNSFFKKPPQRKWTWLSPDGVTRNEIDFIMSTNRQIFNDVSVINSVKTGSDHRIVRGMLNINVRLERSRLMKSTLRPSNAQIQNPESFQLELANRFEWLNSCASVDDVNSRLRRLQSQLTKLKTEDGRVTSNKSEVLREIERFYGQLFTSVSAEPQGLAADPRAKLTRHYTEDIPDISLYEIRMALKQLKNNKAPGEDGITTELLKAGGTPVLKVLQKLFNSSLLDGKPPQAWSRGVVILFFKKGDKTSLKNYRPIALLSHIYKLFSRVITKRLERRFDDFQPTEQAGFRKGYSTIDHIHTLRQIVQKTEEYNRPLCLAFVDYEKAFDSVETWAVLRSLQRCRIDHRYIEVLKCLYNNATMSVRVQEHCTKEIPVKRGVRQGDVISPKLFITALEDSFKLLEWQGLGININDEYITHLRFADDIVVMAESLEDLGRLLGDLSRVSQQVGLKMNMDKTKIMYNVHVAPTIVTVGSSTLEVVDEYIYLGQAVRLGRSNFEREVSRRIQLGWAAFGKLHNVFSSKIPQCLKTKAYNQCVLPVMTYGSETWCFTRDLFRRLKVAQRAMERAMLGISLRDRIRNEKIRRRTKVADIARKISELKWQWAGHIARREDGRWGRKILEWRPRTGKRSVDGHLLDGPTT
Summary
Uniprot
EMBL
FJ265545
ADI61813.1
RSAL01001818
RVE40675.1
AAGJ04050748
MRZV01002136
+ More
PIK34564.1 DS268655 EFO97114.1 JARK01001353 EYC22263.1 JARK01000173 EYC41342.1 JARK01000388 EYC37469.1 JARK01001370 EYC16150.1 EYC16376.1 JARK01001438 EYC02111.1 DS268453 EFP04265.1 JARK01001473 EYB97806.1 JARK01001348 EYC25050.1 EYC25048.1 EYC25046.1 EYC25049.1
PIK34564.1 DS268655 EFO97114.1 JARK01001353 EYC22263.1 JARK01000173 EYC41342.1 JARK01000388 EYC37469.1 JARK01001370 EYC16150.1 EYC16376.1 JARK01001438 EYC02111.1 DS268453 EFP04265.1 JARK01001473 EYB97806.1 JARK01001348 EYC25050.1 EYC25048.1 EYC25046.1 EYC25049.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
PDB
6AR3
E-value=1.95774e-06,
Score=127
Ontologies
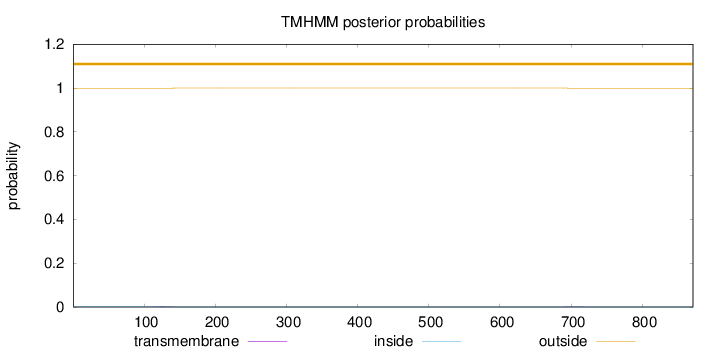
Topology
Length:
871
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04126
Exp number, first 60 AAs:
0.00223
Total prob of N-in:
0.00159
outside
1 - 871
Population Genetic Test Statistics
Pi
361.066548
Theta
216.00396
Tajima's D
2.257594
CLR
0.159505
CSRT
0.91740412979351
Interpretation
Uncertain
