Gene
KWMTBOMO14642
Pre Gene Modal
BGIBMGA009567
Annotation
PREDICTED:_glutamate_receptor_ionotropic?_kainate_3_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.573
Sequence
CDS
ATGAACTTTGACTACGAGATCGTTTCACCGAGGAGCGGGGGATTCGGAAGGAGGCTACCTAATGGCTCCTGGGACGGTGTCGTCGGTGACCTCACTACGGGGGAGACCGACATAGCCGTCGCTGCGTTGACCATGACGGCTGAGAGGGAGGAAGTCATAGATTTCGTGGCGCCATACTTCGAACAGACAGGAATATTGATAGCGATTAGGAAGCCAATACGGAAGACGTCCCTCTTCAAGTTCATGACGGTACTCCGCACTGAGGTTTGGCTCAGCATCGTGGCGGCTCTCGTTCTAACCGGCTTCATGATTTGGCTGCTGGAGAAGTATTCCCCTTACTCGGCGAAGAACAATCCAGGGGCCTACCCATATCCTTGCAGAGATTTTACGCTGAAAGAGAGCTTTTGGTTCGCCCTGACCTCATTCACTCCTCAAGGCGGCGGAGAAGCTCCGAAAGCCCTCTCAGGCAGGACCCTCGTGGCTGCCTACTGGTTATTCGTGGTGCTGATGCTGGCCACATTCACGGCGAACCTTGCTGCTTTCCTCACGGTCGAGAGGATGCAGACTCCGGTGTCATCATTGGAACAGTTGGCGCGTCAATCCCGCATCAATTATACAGTCGTCGAAGGTTCTAGCACGCATCAATACTTCATCAATATGAAGTTCGCCGAAGATACACTATACAGGTATGCATTTTAA
Protein
MNFDYEIVSPRSGGFGRRLPNGSWDGVVGDLTTGETDIAVAALTMTAEREEVIDFVAPYFEQTGILIAIRKPIRKTSLFKFMTVLRTEVWLSIVAALVLTGFMIWLLEKYSPYSAKNNPGAYPYPCRDFTLKESFWFALTSFTPQGGGEAPKALSGRTLVAAYWLFVVLMLATFTANLAAFLTVERMQTPVSSLEQLARQSRINYTVVEGSSTHQYFINMKFAEDTLYRYAF
Summary
Similarity
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family.
Uniprot
H9JJ66
A0A223HD91
A0A2D0WKU4
A0A140G9I7
A0A2D0WKS6
A0A0U3BYI2
+ More
A0A0S1TNU5 A0A076E630 A0A212F0K0 A0A2H1VEU5 A0A386H9C9 A0A146JYM8 A0A0F7QED2 A0A345BF34 A0A2K8GKY0 A0A345BF09 A0A1Q1PP61 A0A076E642 A0A3G6V6M9 A0A223HCW7 H9A5S4 A0A3Q8HDA4 A0A194QB91 A0A0K8TUD1 A0A1B3P5G7 A0A1B0CGT3 A0A1B3B7D3 A0A1A9W8M4 A0A1B0D2N4 A0A1W6L149 A0A0S3J2N6 A0A0T6AU00 A0A1B0GFM5 Q1PQ92 A0A3L9LUK4 A0A2J7RGC2 E2BSR3 E2AH45 A0A026WMY8 M4VR69 A0A310SBH3 N6T0C3 A0A2P8ZDP6 A0A1A9UEK8 A0A1W4XIL7 E0W2Q6 A0A385IUV3 A0A1B0ADQ4 A0A3Q7YRF7 A0A3Q7YR80 A0A3Q7YRA6 A0A3Q7YSK1 D6WHU3 A0A2J7RGC7 A0A1A9XSJ0 A0A1B0AQH3 A0A068F7N4 A0A182SNM6 A0A0N0U733 A0A0C9RXP2 A0A0C9S0B2 A0A088A868 A0A3Q9EL68 A0A182YLZ0 A0A195CUQ6 A0A2P9JY84 A0A348AZW7 A0A0L0CG86 F4WC68 A0A0L7QVT7 A0A158N9Z6 A0A182RRE3 A0A151WH49 A0A182PW52 Q7QDT5 A0A0M4EJA0 A0A182LN75 A0A182MFV8 A0A2A3E8F7 A0A0K8VZJ6 A0A182HX70 A0A182W3Q4 A0A0Q9XH31 B3MZN0 Q17GP1 A0A1I8N2M6 A0A3B0KRS2 A0A182XAZ8 A0A182NDI8 A0A182K301 A0A1W4VGT8 V5MZ71 B4NC58 B5DM21 B4PWU2 A0A0K0PKU4 A0A0N1PH93 A0A182QV01 A0A0M5KAR1 B3NT92
A0A0S1TNU5 A0A076E630 A0A212F0K0 A0A2H1VEU5 A0A386H9C9 A0A146JYM8 A0A0F7QED2 A0A345BF34 A0A2K8GKY0 A0A345BF09 A0A1Q1PP61 A0A076E642 A0A3G6V6M9 A0A223HCW7 H9A5S4 A0A3Q8HDA4 A0A194QB91 A0A0K8TUD1 A0A1B3P5G7 A0A1B0CGT3 A0A1B3B7D3 A0A1A9W8M4 A0A1B0D2N4 A0A1W6L149 A0A0S3J2N6 A0A0T6AU00 A0A1B0GFM5 Q1PQ92 A0A3L9LUK4 A0A2J7RGC2 E2BSR3 E2AH45 A0A026WMY8 M4VR69 A0A310SBH3 N6T0C3 A0A2P8ZDP6 A0A1A9UEK8 A0A1W4XIL7 E0W2Q6 A0A385IUV3 A0A1B0ADQ4 A0A3Q7YRF7 A0A3Q7YR80 A0A3Q7YRA6 A0A3Q7YSK1 D6WHU3 A0A2J7RGC7 A0A1A9XSJ0 A0A1B0AQH3 A0A068F7N4 A0A182SNM6 A0A0N0U733 A0A0C9RXP2 A0A0C9S0B2 A0A088A868 A0A3Q9EL68 A0A182YLZ0 A0A195CUQ6 A0A2P9JY84 A0A348AZW7 A0A0L0CG86 F4WC68 A0A0L7QVT7 A0A158N9Z6 A0A182RRE3 A0A151WH49 A0A182PW52 Q7QDT5 A0A0M4EJA0 A0A182LN75 A0A182MFV8 A0A2A3E8F7 A0A0K8VZJ6 A0A182HX70 A0A182W3Q4 A0A0Q9XH31 B3MZN0 Q17GP1 A0A1I8N2M6 A0A3B0KRS2 A0A182XAZ8 A0A182NDI8 A0A182K301 A0A1W4VGT8 V5MZ71 B4NC58 B5DM21 B4PWU2 A0A0K0PKU4 A0A0N1PH93 A0A182QV01 A0A0M5KAR1 B3NT92
Pubmed
19121390
28150741
27004525
26657286
26812239
24998398
+ More
22118469 25803580 29727827 28736530 30465940 22363688 27006164 26354079 26017144 27538507 27171401 26626891 16520459 20798317 24508170 30249741 23517120 23537049 29403074 20566863 30195211 18362917 19820115 25880816 30542294 25244985 30026095 26108605 21719571 21347285 12364791 14747013 17210077 20966253 17994087 17510324 25315136 15632085 17550304 26265180
22118469 25803580 29727827 28736530 30465940 22363688 27006164 26354079 26017144 27538507 27171401 26626891 16520459 20798317 24508170 30249741 23517120 23537049 29403074 20566863 30195211 18362917 19820115 25880816 30542294 25244985 30026095 26108605 21719571 21347285 12364791 14747013 17210077 20966253 17994087 17510324 25315136 15632085 17550304 26265180
EMBL
BABH01041574
BABH01041575
BABH01041576
KY283697
AST36356.1
KY019180
+ More
ARB05669.1 KU702639 AMM70666.1 KY019176 ARB05665.1 KP975100 ALT31619.1 KR912018 ALM24945.1 KF487713 AII01111.1 AGBW02011068 OWR47276.1 ODYU01001918 SOQ38794.1 MG546652 AYD42275.1 GEDO01000002 JAP88624.1 LC017782 BAR64796.1 MG820687 AXF48858.1 KX585392 ARO70285.1 MG820662 AXF48833.1 KY325448 AQM73610.1 KF487723 AII01121.1 MH193957 AZB49403.1 KY283566 AST36226.1 JN836724 AFC91764.2 MF625612 AXY83439.1 KQ459249 KPJ02260.1 GCVX01000131 JAI18099.1 KX655896 AOG12845.1 AJWK01011573 KU291864 AOE48114.1 AJVK01010713 KX609440 ARN17847.1 KT381532 ALR72538.1 LJIG01022801 KRT78643.1 CCAG010022501 DQ334162 ABC73508.1 KB465734 RLZ02177.1 NEVH01004401 PNF39891.1 GL450241 EFN81309.1 GL439483 EFN67162.1 KK107148 QOIP01000009 EZA57395.1 RLU18233.1 KC113421 AGI05169.1 GGOB01000041 MCH29437.1 APGK01057505 APGK01057506 KB741280 ENN70958.1 PYGN01000087 PSN54615.1 DS235879 EEB19912.1 MH230260 AXY87919.1 GQ316689 ADG54644.1 GQ316659 GQ316660 GQ316662 GQ316663 GQ316664 GQ316665 GQ316666 GQ316667 GQ316668 GQ316669 GQ316670 ADG54614.1 ADG54615.1 ADG54617.1 ADG54618.1 ADG54619.1 ADG54620.1 ADG54621.1 ADG54622.1 ADG54623.1 ADG54624.1 ADG54625.1 GQ316686 GQ316687 GQ316688 GQ316692 GQ316693 ADG54641.1 ADG54642.1 ADG54643.1 ADG54647.1 ADG54648.1 GQ316672 GQ316673 GQ316674 GQ316675 GQ316676 GQ316677 GQ316678 GQ316680 GQ316681 GQ316682 GQ316683 GQ316684 ADG54627.1 ADG54628.1 ADG54629.1 ADG54630.1 ADG54631.1 ADG54632.1 ADG54633.1 ADG54635.1 ADG54636.1 ADG54637.1 ADG54638.1 ADG54639.1 KQ971321 EFA00693.2 PNF39889.1 JXJN01001873 KJ702118 AID61272.1 KQ435720 KOX78574.1 GBYB01013965 JAG83732.1 GBYB01013966 JAG83733.1 MK049043 AZQ24984.1 KQ977276 KYN04416.1 KY817073 AVH87289.1 FX985958 BBD43493.1 JRES01000424 KNC31398.1 GL888070 EGI68048.1 KQ414721 KOC62738.1 ADTU01009924 KQ983135 KYQ47179.1 AAAB01008849 EAA07123.4 CP012528 ALC49166.1 AXCM01001012 KZ288325 PBC28053.1 GDHF01008289 JAI44025.1 APCN01005146 CH933815 KRG07589.1 CH902635 EDV33831.1 CH477257 EAT45826.1 OUUW01000015 SPP88596.1 KF528687 AHA80144.1 CH964239 EDW82417.1 CH379064 EDY72537.1 CM000162 EDX02826.1 KR349063 AKQ25182.1 KQ460253 KPJ16363.1 AXCN02001759 KP843224 ALD51360.1 CH954180 EDV46272.1
ARB05669.1 KU702639 AMM70666.1 KY019176 ARB05665.1 KP975100 ALT31619.1 KR912018 ALM24945.1 KF487713 AII01111.1 AGBW02011068 OWR47276.1 ODYU01001918 SOQ38794.1 MG546652 AYD42275.1 GEDO01000002 JAP88624.1 LC017782 BAR64796.1 MG820687 AXF48858.1 KX585392 ARO70285.1 MG820662 AXF48833.1 KY325448 AQM73610.1 KF487723 AII01121.1 MH193957 AZB49403.1 KY283566 AST36226.1 JN836724 AFC91764.2 MF625612 AXY83439.1 KQ459249 KPJ02260.1 GCVX01000131 JAI18099.1 KX655896 AOG12845.1 AJWK01011573 KU291864 AOE48114.1 AJVK01010713 KX609440 ARN17847.1 KT381532 ALR72538.1 LJIG01022801 KRT78643.1 CCAG010022501 DQ334162 ABC73508.1 KB465734 RLZ02177.1 NEVH01004401 PNF39891.1 GL450241 EFN81309.1 GL439483 EFN67162.1 KK107148 QOIP01000009 EZA57395.1 RLU18233.1 KC113421 AGI05169.1 GGOB01000041 MCH29437.1 APGK01057505 APGK01057506 KB741280 ENN70958.1 PYGN01000087 PSN54615.1 DS235879 EEB19912.1 MH230260 AXY87919.1 GQ316689 ADG54644.1 GQ316659 GQ316660 GQ316662 GQ316663 GQ316664 GQ316665 GQ316666 GQ316667 GQ316668 GQ316669 GQ316670 ADG54614.1 ADG54615.1 ADG54617.1 ADG54618.1 ADG54619.1 ADG54620.1 ADG54621.1 ADG54622.1 ADG54623.1 ADG54624.1 ADG54625.1 GQ316686 GQ316687 GQ316688 GQ316692 GQ316693 ADG54641.1 ADG54642.1 ADG54643.1 ADG54647.1 ADG54648.1 GQ316672 GQ316673 GQ316674 GQ316675 GQ316676 GQ316677 GQ316678 GQ316680 GQ316681 GQ316682 GQ316683 GQ316684 ADG54627.1 ADG54628.1 ADG54629.1 ADG54630.1 ADG54631.1 ADG54632.1 ADG54633.1 ADG54635.1 ADG54636.1 ADG54637.1 ADG54638.1 ADG54639.1 KQ971321 EFA00693.2 PNF39889.1 JXJN01001873 KJ702118 AID61272.1 KQ435720 KOX78574.1 GBYB01013965 JAG83732.1 GBYB01013966 JAG83733.1 MK049043 AZQ24984.1 KQ977276 KYN04416.1 KY817073 AVH87289.1 FX985958 BBD43493.1 JRES01000424 KNC31398.1 GL888070 EGI68048.1 KQ414721 KOC62738.1 ADTU01009924 KQ983135 KYQ47179.1 AAAB01008849 EAA07123.4 CP012528 ALC49166.1 AXCM01001012 KZ288325 PBC28053.1 GDHF01008289 JAI44025.1 APCN01005146 CH933815 KRG07589.1 CH902635 EDV33831.1 CH477257 EAT45826.1 OUUW01000015 SPP88596.1 KF528687 AHA80144.1 CH964239 EDW82417.1 CH379064 EDY72537.1 CM000162 EDX02826.1 KR349063 AKQ25182.1 KQ460253 KPJ16363.1 AXCN02001759 KP843224 ALD51360.1 CH954180 EDV46272.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000092461
UP000091820
UP000092462
+ More
UP000092444 UP000235965 UP000008237 UP000000311 UP000053097 UP000279307 UP000019118 UP000245037 UP000078200 UP000192223 UP000009046 UP000092445 UP000007266 UP000092443 UP000092460 UP000075901 UP000053105 UP000005203 UP000076408 UP000078542 UP000037069 UP000007755 UP000053825 UP000005205 UP000075900 UP000075809 UP000075885 UP000007062 UP000092553 UP000075882 UP000075883 UP000242457 UP000075840 UP000075920 UP000009192 UP000007801 UP000008820 UP000095301 UP000268350 UP000076407 UP000075884 UP000075881 UP000192221 UP000007798 UP000001819 UP000002282 UP000053240 UP000075886 UP000008711
UP000092444 UP000235965 UP000008237 UP000000311 UP000053097 UP000279307 UP000019118 UP000245037 UP000078200 UP000192223 UP000009046 UP000092445 UP000007266 UP000092443 UP000092460 UP000075901 UP000053105 UP000005203 UP000076408 UP000078542 UP000037069 UP000007755 UP000053825 UP000005205 UP000075900 UP000075809 UP000075885 UP000007062 UP000092553 UP000075882 UP000075883 UP000242457 UP000075840 UP000075920 UP000009192 UP000007801 UP000008820 UP000095301 UP000268350 UP000076407 UP000075884 UP000075881 UP000192221 UP000007798 UP000001819 UP000002282 UP000053240 UP000075886 UP000008711
PRIDE
Interpro
SUPFAM
SSF53822
SSF53822
ProteinModelPortal
H9JJ66
A0A223HD91
A0A2D0WKU4
A0A140G9I7
A0A2D0WKS6
A0A0U3BYI2
+ More
A0A0S1TNU5 A0A076E630 A0A212F0K0 A0A2H1VEU5 A0A386H9C9 A0A146JYM8 A0A0F7QED2 A0A345BF34 A0A2K8GKY0 A0A345BF09 A0A1Q1PP61 A0A076E642 A0A3G6V6M9 A0A223HCW7 H9A5S4 A0A3Q8HDA4 A0A194QB91 A0A0K8TUD1 A0A1B3P5G7 A0A1B0CGT3 A0A1B3B7D3 A0A1A9W8M4 A0A1B0D2N4 A0A1W6L149 A0A0S3J2N6 A0A0T6AU00 A0A1B0GFM5 Q1PQ92 A0A3L9LUK4 A0A2J7RGC2 E2BSR3 E2AH45 A0A026WMY8 M4VR69 A0A310SBH3 N6T0C3 A0A2P8ZDP6 A0A1A9UEK8 A0A1W4XIL7 E0W2Q6 A0A385IUV3 A0A1B0ADQ4 A0A3Q7YRF7 A0A3Q7YR80 A0A3Q7YRA6 A0A3Q7YSK1 D6WHU3 A0A2J7RGC7 A0A1A9XSJ0 A0A1B0AQH3 A0A068F7N4 A0A182SNM6 A0A0N0U733 A0A0C9RXP2 A0A0C9S0B2 A0A088A868 A0A3Q9EL68 A0A182YLZ0 A0A195CUQ6 A0A2P9JY84 A0A348AZW7 A0A0L0CG86 F4WC68 A0A0L7QVT7 A0A158N9Z6 A0A182RRE3 A0A151WH49 A0A182PW52 Q7QDT5 A0A0M4EJA0 A0A182LN75 A0A182MFV8 A0A2A3E8F7 A0A0K8VZJ6 A0A182HX70 A0A182W3Q4 A0A0Q9XH31 B3MZN0 Q17GP1 A0A1I8N2M6 A0A3B0KRS2 A0A182XAZ8 A0A182NDI8 A0A182K301 A0A1W4VGT8 V5MZ71 B4NC58 B5DM21 B4PWU2 A0A0K0PKU4 A0A0N1PH93 A0A182QV01 A0A0M5KAR1 B3NT92
A0A0S1TNU5 A0A076E630 A0A212F0K0 A0A2H1VEU5 A0A386H9C9 A0A146JYM8 A0A0F7QED2 A0A345BF34 A0A2K8GKY0 A0A345BF09 A0A1Q1PP61 A0A076E642 A0A3G6V6M9 A0A223HCW7 H9A5S4 A0A3Q8HDA4 A0A194QB91 A0A0K8TUD1 A0A1B3P5G7 A0A1B0CGT3 A0A1B3B7D3 A0A1A9W8M4 A0A1B0D2N4 A0A1W6L149 A0A0S3J2N6 A0A0T6AU00 A0A1B0GFM5 Q1PQ92 A0A3L9LUK4 A0A2J7RGC2 E2BSR3 E2AH45 A0A026WMY8 M4VR69 A0A310SBH3 N6T0C3 A0A2P8ZDP6 A0A1A9UEK8 A0A1W4XIL7 E0W2Q6 A0A385IUV3 A0A1B0ADQ4 A0A3Q7YRF7 A0A3Q7YR80 A0A3Q7YRA6 A0A3Q7YSK1 D6WHU3 A0A2J7RGC7 A0A1A9XSJ0 A0A1B0AQH3 A0A068F7N4 A0A182SNM6 A0A0N0U733 A0A0C9RXP2 A0A0C9S0B2 A0A088A868 A0A3Q9EL68 A0A182YLZ0 A0A195CUQ6 A0A2P9JY84 A0A348AZW7 A0A0L0CG86 F4WC68 A0A0L7QVT7 A0A158N9Z6 A0A182RRE3 A0A151WH49 A0A182PW52 Q7QDT5 A0A0M4EJA0 A0A182LN75 A0A182MFV8 A0A2A3E8F7 A0A0K8VZJ6 A0A182HX70 A0A182W3Q4 A0A0Q9XH31 B3MZN0 Q17GP1 A0A1I8N2M6 A0A3B0KRS2 A0A182XAZ8 A0A182NDI8 A0A182K301 A0A1W4VGT8 V5MZ71 B4NC58 B5DM21 B4PWU2 A0A0K0PKU4 A0A0N1PH93 A0A182QV01 A0A0M5KAR1 B3NT92
PDB
5IDF
E-value=2.63955e-34,
Score=361
Ontologies
GO
GO:0004970
GO:0005886
GO:0016021
GO:0006811
GO:0032281
GO:0004971
GO:0042048
GO:0071683
GO:0010157
GO:0043235
GO:0036082
GO:0019226
GO:0050850
GO:0004984
GO:0005887
GO:0005102
GO:0032590
GO:0071311
GO:0015026
GO:0005234
GO:0016020
GO:0004872
GO:0005216
GO:0003824
GO:0008152
GO:0016491
GO:0015930
GO:0038023
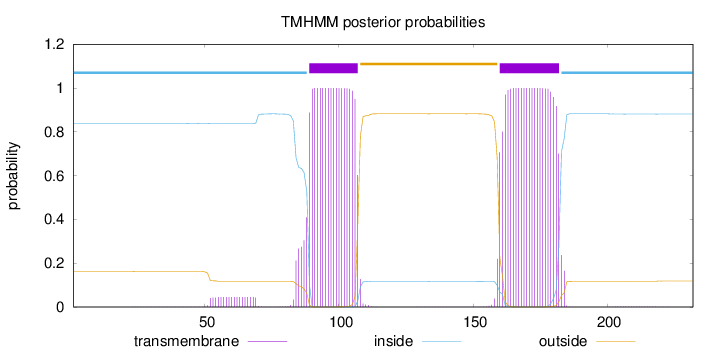
Topology
Length:
232
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.67353
Exp number, first 60 AAs:
0.41804
Total prob of N-in:
0.83739
inside
1 - 88
TMhelix
89 - 107
outside
108 - 159
TMhelix
160 - 182
inside
183 - 232
Population Genetic Test Statistics
Pi
242.170082
Theta
183.749586
Tajima's D
2.151987
CLR
0.549516
CSRT
0.904104794760262
Interpretation
Uncertain
