Gene
KWMTBOMO14631
Pre Gene Modal
BGIBMGA009552
Annotation
PREDICTED:_uncharacterized_protein_LOC106721395_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.495
Sequence
CDS
ATGTCGGTTGGCCTCCGCGGAGTATCTTCGTCCTCAGAGACCATGTCCGAGAGCACGTTAGAGAGAAACCGAGCTGAAAAGGCTCTGATAACAAAAACTCCAACGCATACGTTACTCAAAAGTAATATTAAAAATTATTCGGACAGGAACGGGAACGATGTCACGGAAACCGTACACAGAGGGAGGAAAACCTTCGCATTTTGGACTCTCGTTTGTCTTTTATTCGTATTGGCTATCGGGAACTTAATTCTGACATTCACGATACTAGCCGTCTTGAGATTGGGACAAGGTCTAGAGAGCATGGAGTTTCTTCCGGAACACAATGCTATAAAGTTCTTTGGTAAAACCAACTTGGAGAATGTATATAAGAGAGACGGTTTGATTGAGAGCTTCAGTGATACACCCATGAGCATAACCAGCGAGAATGGCTCTGTTCTATTCAATCTACAGACTAGACTATCACGATCGGACACGAAACTGCTAATGAATTCGTCCGGGGTCTTCATCCGCGGCGTGAACTCGTTGGAGCTGATCAGCCTCGACTCCGGGGAGGCTGCGTTCAGCACAGCATCCCCCGAGATCAATATACACGAGCCGATCAACAATTTGCAGGCGAAAATGGTGAATAAATATAAAAAAAAAACTAAGCAAGGGACGTGGGATGAAAATGCTATGCAAACGGCTATTAATTTGTGCAAAGCTGGAGAATCAATAAAAAGCACAGCTAAAAAGTATGGTTTGGCTTATGCAACCTTGTATAGGCATGTAAAAACCGGCACCGTTGCTTCAAAGCTTGGTCGATTCCGTCCGGTTTTCACTGAAGATCAGGAAATAGAGTTGGTGACATATCTCAAAGATATGGATTCTGTATTTTTTGGCTTGACTCGAGATGAATTCAAAAATTTAGCGTTCATCTACGCCAAGAAAAATAATTTGAACTACCCGAATAACTGGGACAAGTACGAAAAAGCTGGAGATGATTGGCTATCGTCATTTTTGTCTCGTCACAATCAAATATCCCTCAGGACTCCAGAGGCAACATCGGTTGCAAGGGCAAAAGGATTTAATAGACATGAAGTTGGCCGTTTTTATGAAAACCTTGAAGCTTTAACAGTAAAATATGATATTGATGCATCTAGATTATACAACATGGATGAAACTGGTATATCTACAACCACAAATAAGCCGCCTAAAGTGCTATCAACGAAAGGAAAGAAACAAGTGGGCATCATCGCCAGTGCAGAACGTGGTCAATTAACTACTGTCATAGGTTGTTGTAATGCCGCTGGTTCATTCCTCCCACCATTCTTAATTTTCGCCAGAAAAAAGATGCAACCCAGACTACTTGATGGTGCGCCTCCCGGCACTCAAGGAACCTGTACTCCAAATGGTTGGACTTCAGGTGAGGTATTCCTTGATTGGATGCATTTTTTTGTGGAACATGTTCGACCTACCCCTGAGAAAAAAATACTTCTATTATTGGATAACCACGAGAGCCATAAGTATTACCCAGCTTTAGATTACGCCAGCAAGAACAATGTCGTAATTTTATCCTTGGCCCCACACACAACTCATAAAATGCAGCCCATGGATGTAGCGGTATACGGCCCACTAAAAACTTACTTTGAAAGGGAGATAAATTTCTTCCAAAAATCGCACCCAGGACGCATTATAAATCAATACGATGTAGCCAGACTGCTATCACCAGCTTTTCTGAAAAGTGCAGTAGCTCAAAATGCAGTTCATGGATTTGAAAGACCTGGTATTTGGCCAGTGAACAAATATGCTTTTGGTGACGAAGATTATGAGCCAGCCGCTGTTTTTGCAGGCATGTCTAATATGAATATTGGCACAAAAAGTACAAACTCACTAGAAACGATAGATATTCCCAGACCTTCTGTAGACACTTCTTCAACTCCTTCGCCTTCGCTTCTTCAAGAACAGATTCCATCAAGTTCAGTAGATCCTCTATCAGAAATATTTCCATCTCGAAACAGAACTTCCTCTTGTGAGTTTCAAGACTTTGTTGCGCCCATTGTTAACACGCGTGAAAAGTTTCGTAGCGACGGGTGTACACCTGAACCAGAACCTGGATGTTCCCGAATTGTTTCCGCTTTAGAGCCAGATATAGCTGCTCAGAGAGGTGACACAGTGGTGCCAGATATTGAAGTAAATACTCGGCAAAATACTAGCTGTGACAACACATCTGATCCTAAACCGGAATGCTCAGCCTCCCATTGCAGTCCCTTGGTGTTAAGACCTATTCCAAATCCTGCCAAACCAATGACCACGCGAAAACGGAAATTACAAAAATCGGAAATATTAACCAGTACCCCGATCAAAGAAGATCAAAAGCATAAATATGAGAAAAATAAAGTTAAGAATATTAATAAAAGACTGAATGATAAGTCAAAAAATGTTCCTAAGAAGATTAATACAAAGACTGACAAAGACAAAAAAACTACTAAAAAGGTTAAACAGCCAAAGAAAATAAAAGAATTTAAGGATATTTTATGCTTTTTTTGCGGTGAAATATATATAGAAGAAGACGGTAAACCAATTGAAAATTGGATTCAATGTGACATTTGTCAACAATGGTGTCACGAAGATTGTTCTGCTTTTGATGGCGCTCTAGGTTATACCTGTGACAATTGCCAATCTTAA
Protein
MSVGLRGVSSSSETMSESTLERNRAEKALITKTPTHTLLKSNIKNYSDRNGNDVTETVHRGRKTFAFWTLVCLLFVLAIGNLILTFTILAVLRLGQGLESMEFLPEHNAIKFFGKTNLENVYKRDGLIESFSDTPMSITSENGSVLFNLQTRLSRSDTKLLMNSSGVFIRGVNSLELISLDSGEAAFSTASPEINIHEPINNLQAKMVNKYKKKTKQGTWDENAMQTAINLCKAGESIKSTAKKYGLAYATLYRHVKTGTVASKLGRFRPVFTEDQEIELVTYLKDMDSVFFGLTRDEFKNLAFIYAKKNNLNYPNNWDKYEKAGDDWLSSFLSRHNQISLRTPEATSVARAKGFNRHEVGRFYENLEALTVKYDIDASRLYNMDETGISTTTNKPPKVLSTKGKKQVGIIASAERGQLTTVIGCCNAAGSFLPPFLIFARKKMQPRLLDGAPPGTQGTCTPNGWTSGEVFLDWMHFFVEHVRPTPEKKILLLLDNHESHKYYPALDYASKNNVVILSLAPHTTHKMQPMDVAVYGPLKTYFEREINFFQKSHPGRIINQYDVARLLSPAFLKSAVAQNAVHGFERPGIWPVNKYAFGDEDYEPAAVFAGMSNMNIGTKSTNSLETIDIPRPSVDTSSTPSPSLLQEQIPSSSVDPLSEIFPSRNRTSSCEFQDFVAPIVNTREKFRSDGCTPEPEPGCSRIVSALEPDIAAQRGDTVVPDIEVNTRQNTSCDNTSDPKPECSASHCSPLVLRPIPNPAKPMTTRKRKLQKSEILTSTPIKEDQKHKYEKNKVKNINKRLNDKSKNVPKKINTKTDKDKKTTKKVKQPKKIKEFKDILCFFCGEIYIEEDGKPIENWIQCDICQQWCHEDCSAFDGALGYTCDNCQS
Summary
Uniprot
EMBL
NWSH01004485
PCG65052.1
ODYU01004859
SOQ45169.1
AGBW02012433
OWR45073.1
+ More
ODYU01004314 SOQ44057.1 BABH01000686 GEBQ01021595 JAT18382.1 NWSH01003711 PCG65895.1 ODYU01001473 SOQ37710.1 BABH01016912 BABH01009206 NWSH01000827 PCG73986.1 NWSH01002570 PCG67955.1 NWSH01003154 PCG66851.1 NWSH01000193 PCG78522.1
ODYU01004314 SOQ44057.1 BABH01000686 GEBQ01021595 JAT18382.1 NWSH01003711 PCG65895.1 ODYU01001473 SOQ37710.1 BABH01016912 BABH01009206 NWSH01000827 PCG73986.1 NWSH01002570 PCG67955.1 NWSH01003154 PCG66851.1 NWSH01000193 PCG78522.1
Proteomes
Interpro
IPR013083
Znf_RING/FYVE/PHD
+ More
IPR036880 Kunitz_BPTI_sf
IPR009057 Homeobox-like_sf
IPR011011 Znf_FYVE_PHD
IPR004875 DDE_SF_endonuclease_dom
IPR007889 HTH_Psq
IPR001965 Znf_PHD
IPR006600 HTH_CenpB_DNA-bd_dom
IPR019787 Znf_PHD-finger
IPR003599 Ig_sub
IPR036179 Ig-like_dom_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR007110 Ig-like_dom
IPR036880 Kunitz_BPTI_sf
IPR009057 Homeobox-like_sf
IPR011011 Znf_FYVE_PHD
IPR004875 DDE_SF_endonuclease_dom
IPR007889 HTH_Psq
IPR001965 Znf_PHD
IPR006600 HTH_CenpB_DNA-bd_dom
IPR019787 Znf_PHD-finger
IPR003599 Ig_sub
IPR036179 Ig-like_dom_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR007110 Ig-like_dom
Gene 3D
ProteinModelPortal
Ontologies
GO
Topology
Subcellular location
Nucleus
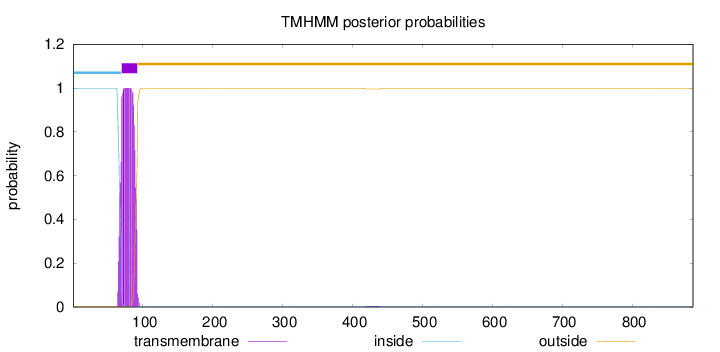
Length:
887
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.31769
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.99876
inside
1 - 69
TMhelix
70 - 92
outside
93 - 887
Population Genetic Test Statistics
Pi
164.511912
Theta
175.517227
Tajima's D
0.442778
CLR
0.611643
CSRT
0.500124993750312
Interpretation
Uncertain
