Pre Gene Modal
BGIBMGA009561
Annotation
PREDICTED:_Kv_channel-interacting_protein_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.103
Sequence
CDS
ATGGCGACGCCTCCCGAGTCCCCAGTCGAAGAACATGCCTACGAATTGGAGCCAAGCAGAACGCCTAAGCCGATACCGGTCGCTTTAGAAGAATTGTGCAGGCAAACTAAATTCTCGAAACAAGAAATTAGAGTTATGTACAGAGGATTTAAAACGGAGTGTCCTGAGGGCGTGGTGCAGGAGGACTCCTTCAAAGATATTTATGCTAAATTCTTTCCACACGGAAATTCAGCCTTATACGCGCACTACGTGTTTAAAGCATTCGACGTGAATTGCAGTGGGGCGATCAGTTTCAGGGAATTACTGGTGACCCTGTCCACGCTGCTCCGTGGCTCCGTGTACGAGCGTCTCCGCTGGGCGTTCCGGCTCTACGACGTGGACGGAGACGGAGCCATCACGAGACAGGAGCTGGCCGAGGTGGTCGTGGCGGTGCACGAGCTTCTGGGCAGACGAGCGCCCCCTGGGTCCCCAGCTGCGAGGCTGGACGATGCCAAAGCCAATGAACAGGTGGACCGGGTGTTCAGGAAGCTGGACCTGAACCAGGACGGCGTGATCACGATCGAGGAGTTCCTTGAGTCGTGTCTGAAGGATGACGCGATCACTAGGTCGCTTCAAATGTTCGACTCGGCACTATGA
Protein
MATPPESPVEEHAYELEPSRTPKPIPVALEELCRQTKFSKQEIRVMYRGFKTECPEGVVQEDSFKDIYAKFFPHGNSALYAHYVFKAFDVNCSGAISFRELLVTLSTLLRGSVYERLRWAFRLYDVDGDGAITRQELAEVVVAVHELLGRRAPPGSPAARLDDAKANEQVDRVFRKLDLNQDGVITIEEFLESCLKDDAITRSLQMFDSAL
Summary
Uniprot
A0A194Q9T6
A0A0N1PHC5
A0A1A9XFJ2
A0A1B0C5H6
A0A1B0CDJ4
A0A1B0FF37
+ More
A0A1A9Z3S2 A0A3B0KM50 A0A0L0CJH1 A0A1I8MU57 A0A1I8PZB4 B4GFX8 Q295N1 D6WA23 B4NJB6 B4JYY5 B4QVF7 B4ICE9 B3P6E2 A0A1W4VLT4 B3LYK6 Q9VBL2 B4PT06 B4K578 B4LYZ2 A0A0M5J5D5 A0A336LQW5 A0A1L8DM47 Q177Q6 A0A182K564 A0A182RPT6 A0A182NCE2 A0A182VZ03 A0A182PKJ6 A0A182XVV6 A0A182MVU4 B0XGS0 A0A182VIH8 A0A182TZU2 A0A182KLS9 A0A182XBC5 Q7PYL8 A0A182I067 A0A182QWJ6 A0A182IT81 A0A084VHG8 W5JI54 A0A182FKU2 J3JV28 A0A1J1IVR1 E0VYH4 N6UP90 A0A067R1S6 A0A3L8DFL5 A0A2H1VTQ5 A0A1B6HLD9 A0A2A3EK47 J9K887 A0A2H8TWD2 A0A2S2PND2 A0A195EVB5 A0A195CLX0 A0A151X555 A0A087ZQS0 A0A336MX74 Q1WWA7 K7J9E4 A0A1Y1N1W1 A0A154PT78 V9IM12 A0A1B6E5J1 A0A226DHC8 E2AAL2 A0A210QRQ4 A0A0L8GY77 A0A3Q4AYM2 A0A3Q3BAV5 A0A3Q3W1S4 A0A3B5MHS2 A0A2G8L355 A0A3Q3GUR5 A0A3B5QMJ9 K1QKC5 A0A2I4BD17 A0A3Q4G2L9 M3ZDC3 A0A3B3YP80 A0A3Q3EV90 A0A0L8GYK1 G3Q2Y7 A0A3Q1K8N3 V4A303 A0A3B3U633 A0A3Q3S2Y4
A0A1A9Z3S2 A0A3B0KM50 A0A0L0CJH1 A0A1I8MU57 A0A1I8PZB4 B4GFX8 Q295N1 D6WA23 B4NJB6 B4JYY5 B4QVF7 B4ICE9 B3P6E2 A0A1W4VLT4 B3LYK6 Q9VBL2 B4PT06 B4K578 B4LYZ2 A0A0M5J5D5 A0A336LQW5 A0A1L8DM47 Q177Q6 A0A182K564 A0A182RPT6 A0A182NCE2 A0A182VZ03 A0A182PKJ6 A0A182XVV6 A0A182MVU4 B0XGS0 A0A182VIH8 A0A182TZU2 A0A182KLS9 A0A182XBC5 Q7PYL8 A0A182I067 A0A182QWJ6 A0A182IT81 A0A084VHG8 W5JI54 A0A182FKU2 J3JV28 A0A1J1IVR1 E0VYH4 N6UP90 A0A067R1S6 A0A3L8DFL5 A0A2H1VTQ5 A0A1B6HLD9 A0A2A3EK47 J9K887 A0A2H8TWD2 A0A2S2PND2 A0A195EVB5 A0A195CLX0 A0A151X555 A0A087ZQS0 A0A336MX74 Q1WWA7 K7J9E4 A0A1Y1N1W1 A0A154PT78 V9IM12 A0A1B6E5J1 A0A226DHC8 E2AAL2 A0A210QRQ4 A0A0L8GY77 A0A3Q4AYM2 A0A3Q3BAV5 A0A3Q3W1S4 A0A3B5MHS2 A0A2G8L355 A0A3Q3GUR5 A0A3B5QMJ9 K1QKC5 A0A2I4BD17 A0A3Q4G2L9 M3ZDC3 A0A3B3YP80 A0A3Q3EV90 A0A0L8GYK1 G3Q2Y7 A0A3Q1K8N3 V4A303 A0A3B3U633 A0A3Q3S2Y4
Pubmed
26354079
26108605
25315136
17994087
15632085
23185243
+ More
18362917 19820115 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17510324 25244985 20966253 12364791 14747013 17210077 24438588 20920257 23761445 22516182 20566863 23537049 24845553 30249741 20075255 28004739 20798317 28812685 29023486 23542700 22992520 25186727 23254933
18362917 19820115 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17510324 25244985 20966253 12364791 14747013 17210077 24438588 20920257 23761445 22516182 20566863 23537049 24845553 30249741 20075255 28004739 20798317 28812685 29023486 23542700 22992520 25186727 23254933
EMBL
KQ459249
KPJ02283.1
KQ460301
KPJ16130.1
JXJN01026003
AJWK01007943
+ More
CCAG010020690 OUUW01000026 SPP89700.1 JRES01000301 KNC32558.1 CH479182 EDW34513.1 CM000070 EAL28677.1 KRT00938.1 KRT00939.1 KRT00940.1 KQ971312 EEZ98073.1 CH964272 EDW84947.1 CH916378 EDV98600.1 CM000364 EDX14483.1 CH480828 EDW45045.1 CH954182 EDV53612.1 CH902617 EDV42921.1 KPU79978.1 KPU79979.1 KPU79980.1 AE014297 BT058099 AAF56519.1 ACM90876.1 CM000160 EDW98693.1 KRK04428.1 CH933806 EDW15078.1 KRG01348.1 KRG01349.1 CH940650 EDW68095.1 KRF83639.1 CP012526 ALC47468.1 UFQS01000043 UFQT01000043 SSW98381.1 SSX18767.1 GFDF01006548 JAV07536.1 CH477372 EAT42408.1 AXCM01011426 AXCM01011427 DS233059 EDS27793.1 AAAB01008987 EAA01229.2 APCN01002050 AXCN02000020 ATLV01013171 KE524842 KFB37412.1 ADMH02001113 ETN64072.1 BT127094 AEE62056.1 CVRI01000062 CRL04192.1 DS235845 EEB18431.1 APGK01023535 KB740498 KB632375 ENN80542.1 ERL93869.1 KK852811 KDR15947.1 QOIP01000009 RLU19081.1 ODYU01004389 SOQ44215.1 GECU01032240 JAS75466.1 KZ288226 PBC31864.1 ABLF02020627 ABLF02020628 ABLF02020631 GFXV01006790 MBW18595.1 GGMR01018296 MBY30915.1 KQ981954 KYN32205.1 KQ977642 KYN01069.1 KQ982519 KYQ55517.1 UFQT01003320 SSX34866.1 BT024999 ABE01229.1 GEZM01015143 JAV91882.1 KQ435207 KZC15073.1 JR050968 AEY61399.1 GEDC01004134 JAS33164.1 LNIX01000020 OXA44107.1 GL438129 EFN69519.1 NEDP02002268 OWF51401.1 KQ419933 KOF81923.1 MRZV01000238 PIK54683.1 JH818480 EKC37182.1 KOF81924.1 KB202284 ESO91082.1
CCAG010020690 OUUW01000026 SPP89700.1 JRES01000301 KNC32558.1 CH479182 EDW34513.1 CM000070 EAL28677.1 KRT00938.1 KRT00939.1 KRT00940.1 KQ971312 EEZ98073.1 CH964272 EDW84947.1 CH916378 EDV98600.1 CM000364 EDX14483.1 CH480828 EDW45045.1 CH954182 EDV53612.1 CH902617 EDV42921.1 KPU79978.1 KPU79979.1 KPU79980.1 AE014297 BT058099 AAF56519.1 ACM90876.1 CM000160 EDW98693.1 KRK04428.1 CH933806 EDW15078.1 KRG01348.1 KRG01349.1 CH940650 EDW68095.1 KRF83639.1 CP012526 ALC47468.1 UFQS01000043 UFQT01000043 SSW98381.1 SSX18767.1 GFDF01006548 JAV07536.1 CH477372 EAT42408.1 AXCM01011426 AXCM01011427 DS233059 EDS27793.1 AAAB01008987 EAA01229.2 APCN01002050 AXCN02000020 ATLV01013171 KE524842 KFB37412.1 ADMH02001113 ETN64072.1 BT127094 AEE62056.1 CVRI01000062 CRL04192.1 DS235845 EEB18431.1 APGK01023535 KB740498 KB632375 ENN80542.1 ERL93869.1 KK852811 KDR15947.1 QOIP01000009 RLU19081.1 ODYU01004389 SOQ44215.1 GECU01032240 JAS75466.1 KZ288226 PBC31864.1 ABLF02020627 ABLF02020628 ABLF02020631 GFXV01006790 MBW18595.1 GGMR01018296 MBY30915.1 KQ981954 KYN32205.1 KQ977642 KYN01069.1 KQ982519 KYQ55517.1 UFQT01003320 SSX34866.1 BT024999 ABE01229.1 GEZM01015143 JAV91882.1 KQ435207 KZC15073.1 JR050968 AEY61399.1 GEDC01004134 JAS33164.1 LNIX01000020 OXA44107.1 GL438129 EFN69519.1 NEDP02002268 OWF51401.1 KQ419933 KOF81923.1 MRZV01000238 PIK54683.1 JH818480 EKC37182.1 KOF81924.1 KB202284 ESO91082.1
Proteomes
UP000053268
UP000053240
UP000092443
UP000092460
UP000092461
UP000092444
+ More
UP000092445 UP000268350 UP000037069 UP000095301 UP000095300 UP000008744 UP000001819 UP000007266 UP000007798 UP000001070 UP000000304 UP000001292 UP000008711 UP000192221 UP000007801 UP000000803 UP000002282 UP000009192 UP000008792 UP000092553 UP000008820 UP000075881 UP000075900 UP000075884 UP000075920 UP000075885 UP000076408 UP000075883 UP000002320 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075886 UP000075880 UP000030765 UP000000673 UP000069272 UP000183832 UP000009046 UP000019118 UP000030742 UP000027135 UP000279307 UP000242457 UP000007819 UP000078541 UP000078542 UP000075809 UP000005203 UP000002358 UP000076502 UP000198287 UP000000311 UP000242188 UP000053454 UP000261620 UP000264800 UP000261380 UP000230750 UP000002852 UP000005408 UP000192220 UP000261580 UP000261480 UP000261660 UP000007635 UP000265040 UP000030746 UP000261500 UP000261640
UP000092445 UP000268350 UP000037069 UP000095301 UP000095300 UP000008744 UP000001819 UP000007266 UP000007798 UP000001070 UP000000304 UP000001292 UP000008711 UP000192221 UP000007801 UP000000803 UP000002282 UP000009192 UP000008792 UP000092553 UP000008820 UP000075881 UP000075900 UP000075884 UP000075920 UP000075885 UP000076408 UP000075883 UP000002320 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075886 UP000075880 UP000030765 UP000000673 UP000069272 UP000183832 UP000009046 UP000019118 UP000030742 UP000027135 UP000279307 UP000242457 UP000007819 UP000078541 UP000078542 UP000075809 UP000005203 UP000002358 UP000076502 UP000198287 UP000000311 UP000242188 UP000053454 UP000261620 UP000264800 UP000261380 UP000230750 UP000002852 UP000005408 UP000192220 UP000261580 UP000261480 UP000261660 UP000007635 UP000265040 UP000030746 UP000261500 UP000261640
Pfam
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194Q9T6
A0A0N1PHC5
A0A1A9XFJ2
A0A1B0C5H6
A0A1B0CDJ4
A0A1B0FF37
+ More
A0A1A9Z3S2 A0A3B0KM50 A0A0L0CJH1 A0A1I8MU57 A0A1I8PZB4 B4GFX8 Q295N1 D6WA23 B4NJB6 B4JYY5 B4QVF7 B4ICE9 B3P6E2 A0A1W4VLT4 B3LYK6 Q9VBL2 B4PT06 B4K578 B4LYZ2 A0A0M5J5D5 A0A336LQW5 A0A1L8DM47 Q177Q6 A0A182K564 A0A182RPT6 A0A182NCE2 A0A182VZ03 A0A182PKJ6 A0A182XVV6 A0A182MVU4 B0XGS0 A0A182VIH8 A0A182TZU2 A0A182KLS9 A0A182XBC5 Q7PYL8 A0A182I067 A0A182QWJ6 A0A182IT81 A0A084VHG8 W5JI54 A0A182FKU2 J3JV28 A0A1J1IVR1 E0VYH4 N6UP90 A0A067R1S6 A0A3L8DFL5 A0A2H1VTQ5 A0A1B6HLD9 A0A2A3EK47 J9K887 A0A2H8TWD2 A0A2S2PND2 A0A195EVB5 A0A195CLX0 A0A151X555 A0A087ZQS0 A0A336MX74 Q1WWA7 K7J9E4 A0A1Y1N1W1 A0A154PT78 V9IM12 A0A1B6E5J1 A0A226DHC8 E2AAL2 A0A210QRQ4 A0A0L8GY77 A0A3Q4AYM2 A0A3Q3BAV5 A0A3Q3W1S4 A0A3B5MHS2 A0A2G8L355 A0A3Q3GUR5 A0A3B5QMJ9 K1QKC5 A0A2I4BD17 A0A3Q4G2L9 M3ZDC3 A0A3B3YP80 A0A3Q3EV90 A0A0L8GYK1 G3Q2Y7 A0A3Q1K8N3 V4A303 A0A3B3U633 A0A3Q3S2Y4
A0A1A9Z3S2 A0A3B0KM50 A0A0L0CJH1 A0A1I8MU57 A0A1I8PZB4 B4GFX8 Q295N1 D6WA23 B4NJB6 B4JYY5 B4QVF7 B4ICE9 B3P6E2 A0A1W4VLT4 B3LYK6 Q9VBL2 B4PT06 B4K578 B4LYZ2 A0A0M5J5D5 A0A336LQW5 A0A1L8DM47 Q177Q6 A0A182K564 A0A182RPT6 A0A182NCE2 A0A182VZ03 A0A182PKJ6 A0A182XVV6 A0A182MVU4 B0XGS0 A0A182VIH8 A0A182TZU2 A0A182KLS9 A0A182XBC5 Q7PYL8 A0A182I067 A0A182QWJ6 A0A182IT81 A0A084VHG8 W5JI54 A0A182FKU2 J3JV28 A0A1J1IVR1 E0VYH4 N6UP90 A0A067R1S6 A0A3L8DFL5 A0A2H1VTQ5 A0A1B6HLD9 A0A2A3EK47 J9K887 A0A2H8TWD2 A0A2S2PND2 A0A195EVB5 A0A195CLX0 A0A151X555 A0A087ZQS0 A0A336MX74 Q1WWA7 K7J9E4 A0A1Y1N1W1 A0A154PT78 V9IM12 A0A1B6E5J1 A0A226DHC8 E2AAL2 A0A210QRQ4 A0A0L8GY77 A0A3Q4AYM2 A0A3Q3BAV5 A0A3Q3W1S4 A0A3B5MHS2 A0A2G8L355 A0A3Q3GUR5 A0A3B5QMJ9 K1QKC5 A0A2I4BD17 A0A3Q4G2L9 M3ZDC3 A0A3B3YP80 A0A3Q3EV90 A0A0L8GYK1 G3Q2Y7 A0A3Q1K8N3 V4A303 A0A3B3U633 A0A3Q3S2Y4
PDB
2NZ0
E-value=8.05791e-49,
Score=485
Ontologies
GO
PANTHER
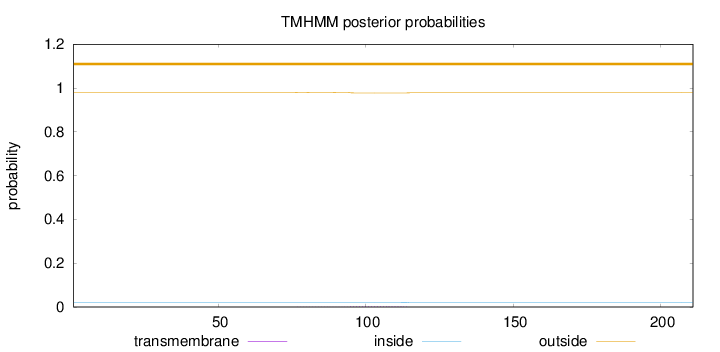
Topology
Length:
211
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0637
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01948
outside
1 - 211
Population Genetic Test Statistics
Pi
202.455265
Theta
173.151654
Tajima's D
1.247506
CLR
0.132726
CSRT
0.730363481825909
Interpretation
Uncertain
