Pre Gene Modal
BGIBMGA009559
Annotation
PREDICTED:_vesicle_transport_protein_GOT1B_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.379
Sequence
CDS
ATGCTTGAGATAACGGATACACAGAAGATTGGCGTAGGCCTAGCTGGTTTCGGAATGACATTTCTATTCCTCGGAGTACTTCTGTTGTTCGATAAAGGTCTCCTAGCCATTGGCAATATCCTGTTCATAAGTGGTCTGACATGTGTGATCGGCATCCAGAGGACGTTTTTCTTCTTCTTCCAACGGCACAAGCTGAAGGCGTCCGTCGCCTTCTTCAGTGGGATCACTATAGTACTGCTCGGCTGGCCCATGATCGGGATGATAGCCGAGATGTACGGCTTCTTGCTGTTGTTCAGAGGATTCCTGCCTTCTGCCATTAATTTCTTAAGGATGGTACCAGTTTTGGGGTCGCTACTGAATCTTCCGATAATAAGAGGGATCGTCGATAGGATAGCGGGAAACAATGGTCGGAACATGGATGACGATCCTTAA
Protein
MLEITDTQKIGVGLAGFGMTFLFLGVLLLFDKGLLAIGNILFISGLTCVIGIQRTFFFFFQRHKLKASVAFFSGITIVLLGWPMIGMIAEMYGFLLLFRGFLPSAINFLRMVPVLGSLLNLPIIRGIVDRIAGNNGRNMDDDP
Summary
Uniprot
H9JJ58
A0A2H1VY29
A0A2A4J2B9
A0A2A4J2D5
A0A3S2LUV1
A0A0N1IPS3
+ More
S4NS88 A0A1S4FRL2 A0A067R0R0 A0A1L8DV02 J9JQD8 A0A1B0CQC7 A0A1B6DV41 A0A1B0D3M9 U5EWB4 A0A2M4C2Z3 A0A139WLM3 A0A023EGA5 T1DFG4 A0A1J1J0L1 B0WFA6 W5JFW1 Q1HQF4 A0A2M3ZL72 A0A1S3DJP2 A0A1Q3G2W1 A0A2M4AMU1 A0A182RCW7 A0A182VQ98 A0A2H8TIU2 A0A182F8N5 A0A2M4C3P9 A0A2R7VXI2 A0A0N7Z9D7 T1HZY7 A0A182N4N4 A0A182QSZ0 A0A0T6BED0 A0A182J8C8 A0A182XIC7 A0A2M3ZD53 A0A2M4AMT0 A0A182PA30 A0A182UME7 A0A182K3Y6 A0A182TSN7 A0A182LFG2 Q7QBY0 A0A182HTB2 A0A0A9X4Z7 J3JWM3 A0A2S2QLG6 E1ZZW9 T1EB25 E9JCP8 A0A154PB23 A0A026VZY8 A0A151J1K1 A0A195CXR9 F4WTW1 A0A195AUL2 A0A151JY00 A0A158P0X6 A0A151WWE2 A0A0K8VIT3 E2BR22 E0W1Z9 B4NEW6 A0A232EPB0 K7IW88 A0A087UNX4 A0A0K8W9W2 A0A0A1WLR8 A0A132A7S3 B3NX67 A0A0J7L8M6 B4IF45 A0A1Y3AW28 A0A1Q3G2P0 B4JJY3 B4MG93 B4PXQ9 A0A131YN96 A0A1I8NT14 A0A023EGG2 B4HB63 A0A1S3DJG2 B4L7Q1 A0A182H1M8 A0A2B4SSW3 A0A087ZUH2 A0A1I8NT02 A0A1I8MLJ3 A0A023GDT5 A7S7J0 A0A1L8ECN0 A0A2A3EJV2 A0A3B0JWZ4 A0A1E1WXE5 A0A023FWG0
S4NS88 A0A1S4FRL2 A0A067R0R0 A0A1L8DV02 J9JQD8 A0A1B0CQC7 A0A1B6DV41 A0A1B0D3M9 U5EWB4 A0A2M4C2Z3 A0A139WLM3 A0A023EGA5 T1DFG4 A0A1J1J0L1 B0WFA6 W5JFW1 Q1HQF4 A0A2M3ZL72 A0A1S3DJP2 A0A1Q3G2W1 A0A2M4AMU1 A0A182RCW7 A0A182VQ98 A0A2H8TIU2 A0A182F8N5 A0A2M4C3P9 A0A2R7VXI2 A0A0N7Z9D7 T1HZY7 A0A182N4N4 A0A182QSZ0 A0A0T6BED0 A0A182J8C8 A0A182XIC7 A0A2M3ZD53 A0A2M4AMT0 A0A182PA30 A0A182UME7 A0A182K3Y6 A0A182TSN7 A0A182LFG2 Q7QBY0 A0A182HTB2 A0A0A9X4Z7 J3JWM3 A0A2S2QLG6 E1ZZW9 T1EB25 E9JCP8 A0A154PB23 A0A026VZY8 A0A151J1K1 A0A195CXR9 F4WTW1 A0A195AUL2 A0A151JY00 A0A158P0X6 A0A151WWE2 A0A0K8VIT3 E2BR22 E0W1Z9 B4NEW6 A0A232EPB0 K7IW88 A0A087UNX4 A0A0K8W9W2 A0A0A1WLR8 A0A132A7S3 B3NX67 A0A0J7L8M6 B4IF45 A0A1Y3AW28 A0A1Q3G2P0 B4JJY3 B4MG93 B4PXQ9 A0A131YN96 A0A1I8NT14 A0A023EGG2 B4HB63 A0A1S3DJG2 B4L7Q1 A0A182H1M8 A0A2B4SSW3 A0A087ZUH2 A0A1I8NT02 A0A1I8MLJ3 A0A023GDT5 A7S7J0 A0A1L8ECN0 A0A2A3EJV2 A0A3B0JWZ4 A0A1E1WXE5 A0A023FWG0
Pubmed
19121390
26354079
23622113
24845553
18362917
19820115
+ More
24945155 24330624 20920257 23761445 17204158 17510324 27129103 20966253 12364791 14747013 17210077 25401762 26823975 22516182 23537049 20798317 21282665 24508170 30249741 21719571 21347285 20566863 17994087 28648823 20075255 25830018 26555130 17550304 26830274 18057021 26483478 25315136 17615350 28503490
24945155 24330624 20920257 23761445 17204158 17510324 27129103 20966253 12364791 14747013 17210077 25401762 26823975 22516182 23537049 20798317 21282665 24508170 30249741 21719571 21347285 20566863 17994087 28648823 20075255 25830018 26555130 17550304 26830274 18057021 26483478 25315136 17615350 28503490
EMBL
BABH01040466
ODYU01005155
SOQ45739.1
NWSH01004041
PCG65542.1
PCG65543.1
+ More
RSAL01000199 RVE44504.1 KQ460301 KPJ16131.1 GAIX01014207 JAA78353.1 KK853024 KDR12335.1 GFDF01003835 JAV10249.1 ABLF02024979 AJWK01023411 GEDC01007742 JAS29556.1 AJVK01003062 GANO01001495 JAB58376.1 GGFJ01010390 MBW59531.1 KQ971321 KYB28793.1 GAPW01005743 GEHC01000353 JAC07855.1 JAV47292.1 GALA01000617 JAA94235.1 CVRI01000066 CRL05872.1 DS231916 EDS26123.1 ADMH02001618 ETN61770.1 DQ440490 CH477701 ABF18523.1 EAT37078.1 EAT37079.1 GGFM01008491 MBW29242.1 GFDL01000949 JAV34096.1 GGFK01008795 MBW42116.1 GFXV01001393 MBW13198.1 GGFJ01010487 MBW59628.1 KK854150 PTY12207.1 GDKW01000835 JAI55760.1 ACPB03008739 AXCN02000052 LJIG01001288 KRT85652.1 GGFM01005736 MBW26487.1 GGFK01008785 MBW42106.1 AAAB01008859 EAA08222.3 APCN01000435 GBHO01031469 GDHC01004193 JAG12135.1 JAQ14436.1 BT127641 KB632070 AEE62603.1 ERL88532.1 GGMS01009406 MBY78609.1 GL435530 EFN73280.1 GAMD01000438 JAB01153.1 GL771854 EFZ09401.1 KQ434846 KZC08428.1 KK107622 QOIP01000009 EZA48444.1 RLU18241.1 KQ980514 KYN15755.1 KQ977141 KYN05455.1 GL888345 EGI62318.1 KQ976738 KYM75747.1 KQ981515 KYN40779.1 ADTU01005894 KQ982691 KYQ52136.1 GDHF01013513 JAI38801.1 GL449898 EFN81828.1 DS235873 EEB19593.1 CH964239 EDW82285.1 NNAY01002956 OXU20219.1 KK120802 KFM79063.1 GDHF01012695 GDHF01004368 JAI39619.1 JAI47946.1 GBXI01014293 JAC99998.1 JXLN01011210 KPM06977.1 CH954180 EDV46896.1 LBMM01000237 KMR03372.1 CH480832 EDW46099.1 MUJZ01055020 OTF72701.1 GFDL01000958 JAV34087.1 CH916370 EDV99885.1 CH940672 EDW57416.1 CM000162 EDX01895.1 GEDV01008130 JAP80427.1 GAPW01005854 JAC07744.1 CH479248 EDW37875.1 CH933814 EDW05764.1 KRG07517.1 JXUM01103695 KQ564826 KXJ71696.1 LSMT01000028 PFX31990.1 GBBM01003416 JAC32002.1 DS469593 EDO40339.1 GFDG01002450 JAV16349.1 KZ288223 PBC31970.1 OUUW01000011 SPP86607.1 GFAC01007478 JAT91710.1 GBBL01002252 JAC25068.1
RSAL01000199 RVE44504.1 KQ460301 KPJ16131.1 GAIX01014207 JAA78353.1 KK853024 KDR12335.1 GFDF01003835 JAV10249.1 ABLF02024979 AJWK01023411 GEDC01007742 JAS29556.1 AJVK01003062 GANO01001495 JAB58376.1 GGFJ01010390 MBW59531.1 KQ971321 KYB28793.1 GAPW01005743 GEHC01000353 JAC07855.1 JAV47292.1 GALA01000617 JAA94235.1 CVRI01000066 CRL05872.1 DS231916 EDS26123.1 ADMH02001618 ETN61770.1 DQ440490 CH477701 ABF18523.1 EAT37078.1 EAT37079.1 GGFM01008491 MBW29242.1 GFDL01000949 JAV34096.1 GGFK01008795 MBW42116.1 GFXV01001393 MBW13198.1 GGFJ01010487 MBW59628.1 KK854150 PTY12207.1 GDKW01000835 JAI55760.1 ACPB03008739 AXCN02000052 LJIG01001288 KRT85652.1 GGFM01005736 MBW26487.1 GGFK01008785 MBW42106.1 AAAB01008859 EAA08222.3 APCN01000435 GBHO01031469 GDHC01004193 JAG12135.1 JAQ14436.1 BT127641 KB632070 AEE62603.1 ERL88532.1 GGMS01009406 MBY78609.1 GL435530 EFN73280.1 GAMD01000438 JAB01153.1 GL771854 EFZ09401.1 KQ434846 KZC08428.1 KK107622 QOIP01000009 EZA48444.1 RLU18241.1 KQ980514 KYN15755.1 KQ977141 KYN05455.1 GL888345 EGI62318.1 KQ976738 KYM75747.1 KQ981515 KYN40779.1 ADTU01005894 KQ982691 KYQ52136.1 GDHF01013513 JAI38801.1 GL449898 EFN81828.1 DS235873 EEB19593.1 CH964239 EDW82285.1 NNAY01002956 OXU20219.1 KK120802 KFM79063.1 GDHF01012695 GDHF01004368 JAI39619.1 JAI47946.1 GBXI01014293 JAC99998.1 JXLN01011210 KPM06977.1 CH954180 EDV46896.1 LBMM01000237 KMR03372.1 CH480832 EDW46099.1 MUJZ01055020 OTF72701.1 GFDL01000958 JAV34087.1 CH916370 EDV99885.1 CH940672 EDW57416.1 CM000162 EDX01895.1 GEDV01008130 JAP80427.1 GAPW01005854 JAC07744.1 CH479248 EDW37875.1 CH933814 EDW05764.1 KRG07517.1 JXUM01103695 KQ564826 KXJ71696.1 LSMT01000028 PFX31990.1 GBBM01003416 JAC32002.1 DS469593 EDO40339.1 GFDG01002450 JAV16349.1 KZ288223 PBC31970.1 OUUW01000011 SPP86607.1 GFAC01007478 JAT91710.1 GBBL01002252 JAC25068.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000027135
UP000007819
+ More
UP000092461 UP000092462 UP000007266 UP000183832 UP000002320 UP000000673 UP000008820 UP000079169 UP000075900 UP000075920 UP000069272 UP000015103 UP000075884 UP000075886 UP000075880 UP000076407 UP000075885 UP000075903 UP000075881 UP000075902 UP000075882 UP000007062 UP000075840 UP000030742 UP000000311 UP000076502 UP000053097 UP000279307 UP000078492 UP000078542 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000008237 UP000009046 UP000007798 UP000215335 UP000002358 UP000054359 UP000008711 UP000036403 UP000001292 UP000001070 UP000008792 UP000002282 UP000095300 UP000008744 UP000009192 UP000069940 UP000249989 UP000225706 UP000005203 UP000095301 UP000001593 UP000242457 UP000268350
UP000092461 UP000092462 UP000007266 UP000183832 UP000002320 UP000000673 UP000008820 UP000079169 UP000075900 UP000075920 UP000069272 UP000015103 UP000075884 UP000075886 UP000075880 UP000076407 UP000075885 UP000075903 UP000075881 UP000075902 UP000075882 UP000007062 UP000075840 UP000030742 UP000000311 UP000076502 UP000053097 UP000279307 UP000078492 UP000078542 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000008237 UP000009046 UP000007798 UP000215335 UP000002358 UP000054359 UP000008711 UP000036403 UP000001292 UP000001070 UP000008792 UP000002282 UP000095300 UP000008744 UP000009192 UP000069940 UP000249989 UP000225706 UP000005203 UP000095301 UP000001593 UP000242457 UP000268350
PRIDE
Pfam
PF04178 Got1
Interpro
IPR007305
Vesicle_transpt_Got1/SFT2
ProteinModelPortal
H9JJ58
A0A2H1VY29
A0A2A4J2B9
A0A2A4J2D5
A0A3S2LUV1
A0A0N1IPS3
+ More
S4NS88 A0A1S4FRL2 A0A067R0R0 A0A1L8DV02 J9JQD8 A0A1B0CQC7 A0A1B6DV41 A0A1B0D3M9 U5EWB4 A0A2M4C2Z3 A0A139WLM3 A0A023EGA5 T1DFG4 A0A1J1J0L1 B0WFA6 W5JFW1 Q1HQF4 A0A2M3ZL72 A0A1S3DJP2 A0A1Q3G2W1 A0A2M4AMU1 A0A182RCW7 A0A182VQ98 A0A2H8TIU2 A0A182F8N5 A0A2M4C3P9 A0A2R7VXI2 A0A0N7Z9D7 T1HZY7 A0A182N4N4 A0A182QSZ0 A0A0T6BED0 A0A182J8C8 A0A182XIC7 A0A2M3ZD53 A0A2M4AMT0 A0A182PA30 A0A182UME7 A0A182K3Y6 A0A182TSN7 A0A182LFG2 Q7QBY0 A0A182HTB2 A0A0A9X4Z7 J3JWM3 A0A2S2QLG6 E1ZZW9 T1EB25 E9JCP8 A0A154PB23 A0A026VZY8 A0A151J1K1 A0A195CXR9 F4WTW1 A0A195AUL2 A0A151JY00 A0A158P0X6 A0A151WWE2 A0A0K8VIT3 E2BR22 E0W1Z9 B4NEW6 A0A232EPB0 K7IW88 A0A087UNX4 A0A0K8W9W2 A0A0A1WLR8 A0A132A7S3 B3NX67 A0A0J7L8M6 B4IF45 A0A1Y3AW28 A0A1Q3G2P0 B4JJY3 B4MG93 B4PXQ9 A0A131YN96 A0A1I8NT14 A0A023EGG2 B4HB63 A0A1S3DJG2 B4L7Q1 A0A182H1M8 A0A2B4SSW3 A0A087ZUH2 A0A1I8NT02 A0A1I8MLJ3 A0A023GDT5 A7S7J0 A0A1L8ECN0 A0A2A3EJV2 A0A3B0JWZ4 A0A1E1WXE5 A0A023FWG0
S4NS88 A0A1S4FRL2 A0A067R0R0 A0A1L8DV02 J9JQD8 A0A1B0CQC7 A0A1B6DV41 A0A1B0D3M9 U5EWB4 A0A2M4C2Z3 A0A139WLM3 A0A023EGA5 T1DFG4 A0A1J1J0L1 B0WFA6 W5JFW1 Q1HQF4 A0A2M3ZL72 A0A1S3DJP2 A0A1Q3G2W1 A0A2M4AMU1 A0A182RCW7 A0A182VQ98 A0A2H8TIU2 A0A182F8N5 A0A2M4C3P9 A0A2R7VXI2 A0A0N7Z9D7 T1HZY7 A0A182N4N4 A0A182QSZ0 A0A0T6BED0 A0A182J8C8 A0A182XIC7 A0A2M3ZD53 A0A2M4AMT0 A0A182PA30 A0A182UME7 A0A182K3Y6 A0A182TSN7 A0A182LFG2 Q7QBY0 A0A182HTB2 A0A0A9X4Z7 J3JWM3 A0A2S2QLG6 E1ZZW9 T1EB25 E9JCP8 A0A154PB23 A0A026VZY8 A0A151J1K1 A0A195CXR9 F4WTW1 A0A195AUL2 A0A151JY00 A0A158P0X6 A0A151WWE2 A0A0K8VIT3 E2BR22 E0W1Z9 B4NEW6 A0A232EPB0 K7IW88 A0A087UNX4 A0A0K8W9W2 A0A0A1WLR8 A0A132A7S3 B3NX67 A0A0J7L8M6 B4IF45 A0A1Y3AW28 A0A1Q3G2P0 B4JJY3 B4MG93 B4PXQ9 A0A131YN96 A0A1I8NT14 A0A023EGG2 B4HB63 A0A1S3DJG2 B4L7Q1 A0A182H1M8 A0A2B4SSW3 A0A087ZUH2 A0A1I8NT02 A0A1I8MLJ3 A0A023GDT5 A7S7J0 A0A1L8ECN0 A0A2A3EJV2 A0A3B0JWZ4 A0A1E1WXE5 A0A023FWG0
Ontologies
GO
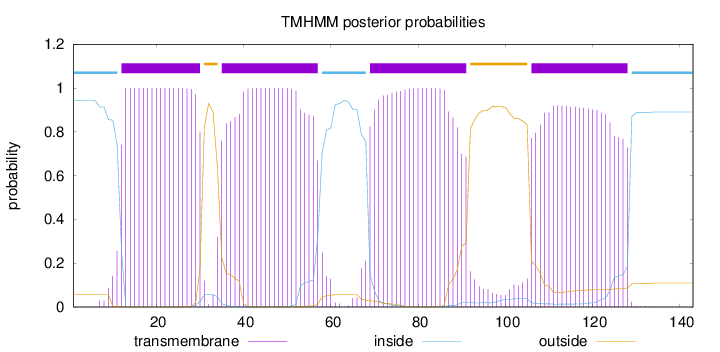
Topology
Length:
143
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
84.65011
Exp number, first 60 AAs:
41.43504
Total prob of N-in:
0.94286
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 30
outside
31 - 34
TMhelix
35 - 57
inside
58 - 68
TMhelix
69 - 91
outside
92 - 105
TMhelix
106 - 128
inside
129 - 143
Population Genetic Test Statistics
Pi
289.546928
Theta
174.339229
Tajima's D
2.244655
CLR
0.143003
CSRT
0.91850407479626
Interpretation
Uncertain
