Gene
KWMTBOMO14621
Pre Gene Modal
BGIBMGA009555
Annotation
PREDICTED:_tankyrase-2_[Amyelois_transitella]
Full name
Poly [ADP-ribose] polymerase
+ More
Poly [ADP-ribose] polymerase tankyrase-1
Poly [ADP-ribose] polymerase tankyrase-1
Alternative Name
ADP-ribosyltransferase diphtheria toxin-like 5
Protein poly-ADP-ribosyltransferase tankyrase-1
TRF1-interacting ankyrin-related ADP-ribose polymerase 1
Tankyrase-1
Protein poly-ADP-ribosyltransferase tankyrase-1
TRF1-interacting ankyrin-related ADP-ribose polymerase 1
Tankyrase-1
Location in the cell
Mitochondrial Reliability : 1.925 Nuclear Reliability : 1.283
Sequence
CDS
ATGGCGGAGAATTTTAGGTCTGCCGCAGATGCTTGCACTAACGCGGCGGCAGTCCGGCTGCTAGAAGCTGCTCGCACTGGTGACGTGGAGACAGCCCGAGCGATCCTCGACTCGCGACCGAGACTCGTCAACTGCAGAGATGTCGACGGGCGGCATTCGACGCCCCTGCATTTCGCTGCCGGGTACAATCGACTTCCATTGGCTCAGTTACTACTTCAGCGTGGAGCTGATGTACACGCGAAAGATAAGGGAGGTCTGGTCCCGCTTCACAACGCGTGCTCCTACGGCCATTACGAGGTCACCGAGCTCCTCGTGTCTGCCGGAGCCGGTGTGAACGCCGCGGACCTCTGGAGGTTCTCACCCTTGCACGAGGCAGCGGCTAAAGGCAAGGCGGATATCGTGAGATTGCTGCTCAAGCATGGGGCGGATCCCACCCGGAGGAACAGGGACGGACTCACGCCGCTGCAACTGGTGCGAGCGGGAGACAGCGACACGGCTGACGCGCTGCGGGGTGATGCTGCGCTACTGGACGCTGCCAAGAGAGGGGACCTCGCTCGAGCCAGGAAACTGATAACTCCTCAAAACGTGAACTGCCGGGACGCGCACGGACGTAACTCCACGCCCTTGCATTTGGCTGCCGGGTATAACAACCTGGAGGTGGCCGAGGCTCTGCTGGAGGCGGGCGCGCTGGTGTCGGCGCGCGACAAGGGCGGGCTGGTGCCGCTGCACAATGCGGCCTCATACGGGCACCTCGAGCTGGCGGCGCTGCTGCTGCGCGCGGGCACGCCGCCCAACGCCGCCGACCGCTGGGGCTTCACGCCCCTGCACGAGGCCGCGCACAAGGCCAGGACCCAGCTCTGCGCGCTGCTGCTGGCCCACGGAGCGGACCCGTTCCTCAAGAACCAGGAGGGGCAGACGGCTCTTGAACTGTCCGGGGCCGAGGACGTGAGGTCGCTGCTGCAGGACGCGATGACGTCATCGGCCGGTGACGTGACGTCATCGTCCGCGCCCGTCCCTCCGCCGGCTCCGCTGCCGCCCCCCTCGCACCCCGTGCTCATGCCGAGCGGGGACACGGTGCAGTTGGCGTTGCCGGTCCTGCCCAGCAACTACTGGGCGGAGGGGTCCAGGGGCAGCTTGGAAGACGCAGCCGGGAGGCCGGCGTCGGCCCTCTCCACCTCGGAGTCCCTGCAGACATTCCTGGCGAGCGTCGGACTGGAGCAGCTGGGGCCGCTGCTGGAGCGAGAGCAGATAACGGTGGACATCCTGTGCGAGATGTCGCACGAGGATCTGCGCGCGGTCGGCGTTGCGGCCTACGGACATCGGCATCGGTTGCTAAAGGCGGCCCGGCGCACCGCCCACGCCGCCAACGAATGGTACGGCGGCACGACCCTCGTAGAGGTGACCCCGAGCGGCGAGGGGGGCGAGTACGCGGTGCTCGAGAGGGAGATGCAGGCCTCCGTGCGCGCGCACCGGGAGCACGCCGGGGGCCACTTCACGCACTACAACGTCGTCAAGATCCAGAAGATCGTGAACGGTCGGGTCTGGGAGCGGTACCAGCACCGGCGGCGCGAGGTCGCAGAGGAGGCCGGCGGCGACAACGAGCGCATGCTCTTCCACGGCTCGCCCTTCATCAACGCCATCGTGCAGAAGGGCTTCGACGAGAGGCACGCCTACATCGGGGGCATGTTCGGCGCCGGGATATACTTCGCGGAGCACTCCTCGAAGAGCAACCAGTACGTGTACGGGTTCGGCGGTGGCTCGGGCTGCCCCGCGCACAAGGACCGGAGCTGCTACCTCTGCCACAGACAGATGCTGCTGTGCCGCGTGACCCTGGGCCGCGGGTTCCAGCTGCTGAGCGCGATGAAGATGGCGCACGCGCCGCCCGGGCACCACTCGGTGGCGGGCCGGCCCTCGCAGGGGGGGCTCTGCTTTCCGGAGTACGTGGTCTACAGGGGGGAGCAGGCCTACCCCGAGTACCTGATCACGTATCAGATCGTGAGCAGCGAGAGCGGCCCGGGCGAGGTGTGA
Protein
MAENFRSAADACTNAAAVRLLEAARTGDVETARAILDSRPRLVNCRDVDGRHSTPLHFAAGYNRLPLAQLLLQRGADVHAKDKGGLVPLHNACSYGHYEVTELLVSAGAGVNAADLWRFSPLHEAAAKGKADIVRLLLKHGADPTRRNRDGLTPLQLVRAGDSDTADALRGDAALLDAAKRGDLARARKLITPQNVNCRDAHGRNSTPLHLAAGYNNLEVAEALLEAGALVSARDKGGLVPLHNAASYGHLELAALLLRAGTPPNAADRWGFTPLHEAAHKARTQLCALLLAHGADPFLKNQEGQTALELSGAEDVRSLLQDAMTSSAGDVTSSSAPVPPPAPLPPPSHPVLMPSGDTVQLALPVLPSNYWAEGSRGSLEDAAGRPASALSTSESLQTFLASVGLEQLGPLLEREQITVDILCEMSHEDLRAVGVAAYGHRHRLLKAARRTAHAANEWYGGTTLVEVTPSGEGGEYAVLEREMQASVRAHREHAGGHFTHYNVVKIQKIVNGRVWERYQHRRREVAEEAGGDNERMLFHGSPFINAIVQKGFDERHAYIGGMFGAGIYFAEHSSKSNQYVYGFGGGSGCPAHKDRSCYLCHRQMLLCRVTLGRGFQLLSAMKMAHAPPGHHSVAGRPSQGGLCFPEYVVYRGEQAYPEYLITYQIVSSESGPGEV
Summary
Description
Poly-ADP-ribosyltransferase involved in various processes such as Wnt signaling pathway, telomere length and vesicle trafficking. Acts as an activator of the Wnt signaling pathway by mediating poly-ADP-ribosylation (PARsylation) of AXIN1 and AXIN2, 2 key components of the beta-catenin destruction complex: poly-ADP-ribosylated target proteins are recognized by RNF146, which mediates their ubiquitination and subsequent degradation. Also mediates PARsylation of BLZF1 and CASC3, followed by recruitment of RNF146 and subsequent ubiquitination. Mediates PARsylation of TERF1, thereby contributing to the regulation of telomere length. Involved in centrosome maturation during prometaphase by mediating PARsylation of HEPACAM2/MIKI. May also regulate vesicle trafficking and modulate the subcellular distribution of SLC2A4/GLUT4-vesicles. May be involved in spindle pole assembly through PARsylation of NUMA1. Stimulates 26S proteasome activity.
Catalytic Activity
NAD(+) + (ADP-D-ribosyl)(n)-acceptor = nicotinamide + (ADP-D-ribosyl)(n+1)-acceptor.
L-aspartyl-[protein] + NAD(+) = 4-O-(ADP-D-ribosyl)-L-aspartyl-[protein] + nicotinamide
L-glutamyl-[protein] + NAD(+) = 5-O-(ADP-D-ribosyl)-L-glutamyl-[protein] + nicotinamide
L-aspartyl-[protein] + NAD(+) = 4-O-(ADP-D-ribosyl)-L-aspartyl-[protein] + nicotinamide
L-glutamyl-[protein] + NAD(+) = 5-O-(ADP-D-ribosyl)-L-glutamyl-[protein] + nicotinamide
Subunit
Oligomerizes and associates with TNKS2. Interacts with the cytoplasmic domain of LNPEP/Otase in SLC2A4/GLUT4-vesicles. Binds to the N-terminus of telomeric TERF1 via the ANK repeats. Found in a complex with POT1; TERF1 and TINF2. Interacts with AXIN1. Interacts with AXIN2. Interacts with BLZF1 and CASC3. Interacts with NUMA1.
Keywords
3D-structure
ADP-ribosylation
Alternative splicing
ANK repeat
Cell cycle
Cell division
Chromosome
Complete proteome
Cytoplasm
Cytoskeleton
Glycosyltransferase
Golgi apparatus
Membrane
Metal-binding
Mitosis
mRNA transport
NAD
Nuclear pore complex
Nucleus
Phosphoprotein
Protein transport
Reference proteome
Repeat
Telomere
Transferase
Translocation
Transport
Ubl conjugation
Wnt signaling pathway
Zinc
Feature
chain Poly [ADP-ribose] polymerase
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JJ54
A0A1Y1MGQ1
D6WBK7
Q17FK9
A0A2J7R2K4
A0A067RRJ4
+ More
A0A087UUU1 E2A4P2 A0A026W9G4 E9IFR3 A0A151ICA6 F4W9V3 A0A158NZ41 A0A151WKM1 A0A151JP23 A0A0C9QYT9 A0A195BRD1 A0A1Q3G2S5 A0A182YG88 A0A336MPT4 A0A0K8T3I0 A0A182PQV5 A0A0C9R2U6 A0A146ME76 A0A182J3K7 A0A2M4BAR1 A0A2M4BAB1 A0A182MSV2 A0A2M4CUA0 A0A154P3V6 A0A1Y9IS44 A0A2M4AGI6 A0A2M4D0F1 A0A2M4CUJ3 W5JE54 A0A2M4CPG4 A0A336MN10 Q7PYH8 A0A2C9GRX3 T1I1W7 A0A1B6DFS8 A0A1L8DJ06 K1QV17 A0A1L8DIT8 A0A023F473 K7IPE9 A0A0K2UZT5 A0A1B0GJW3 W8BL91 L7LZZ4 A0A232FGN2 A0A2A3EDN9 N6TLM3 A0A088ADB5 A0A210QC45 V5HT76 I6L4N1 A0A0L8GA82 A0A0L0CBM5 A0A1A8FWD1 A0A182VR25 A0A3B3IBB7 A0A1I8PA53 H2TPE9 H2TPF0 H3DNG1 A0A3B3HQ19 A0A3B3I5D9 A0A060X381 L5LS37 A0A182N620 A0A1A9UTL6 S7P8B6 A0A182RXK5 A0A2S2PK85 A0A3P4NNU1 A0A182HBN8 A0A2C9JDF7 A0A384BQD6 Q6PFX9-2 G1N930 A0A1A8HT28 G1LHI1 A0A3Q2I900 A0A182F895 A0A0P7XKI1 A0A2K6NKJ4 A0A1A8PSE2 A0A147A1E8 A0A3L7IJA5 A0A341D1L1 A0A2Y9N107 A0A146QX23 A0A146WDV9 A0A1A8L606 V9KDB3 A0A1A8RV89 G3H624 A0A2K6M6H4 W5PKC3
A0A087UUU1 E2A4P2 A0A026W9G4 E9IFR3 A0A151ICA6 F4W9V3 A0A158NZ41 A0A151WKM1 A0A151JP23 A0A0C9QYT9 A0A195BRD1 A0A1Q3G2S5 A0A182YG88 A0A336MPT4 A0A0K8T3I0 A0A182PQV5 A0A0C9R2U6 A0A146ME76 A0A182J3K7 A0A2M4BAR1 A0A2M4BAB1 A0A182MSV2 A0A2M4CUA0 A0A154P3V6 A0A1Y9IS44 A0A2M4AGI6 A0A2M4D0F1 A0A2M4CUJ3 W5JE54 A0A2M4CPG4 A0A336MN10 Q7PYH8 A0A2C9GRX3 T1I1W7 A0A1B6DFS8 A0A1L8DJ06 K1QV17 A0A1L8DIT8 A0A023F473 K7IPE9 A0A0K2UZT5 A0A1B0GJW3 W8BL91 L7LZZ4 A0A232FGN2 A0A2A3EDN9 N6TLM3 A0A088ADB5 A0A210QC45 V5HT76 I6L4N1 A0A0L8GA82 A0A0L0CBM5 A0A1A8FWD1 A0A182VR25 A0A3B3IBB7 A0A1I8PA53 H2TPE9 H2TPF0 H3DNG1 A0A3B3HQ19 A0A3B3I5D9 A0A060X381 L5LS37 A0A182N620 A0A1A9UTL6 S7P8B6 A0A182RXK5 A0A2S2PK85 A0A3P4NNU1 A0A182HBN8 A0A2C9JDF7 A0A384BQD6 Q6PFX9-2 G1N930 A0A1A8HT28 G1LHI1 A0A3Q2I900 A0A182F895 A0A0P7XKI1 A0A2K6NKJ4 A0A1A8PSE2 A0A147A1E8 A0A3L7IJA5 A0A341D1L1 A0A2Y9N107 A0A146QX23 A0A146WDV9 A0A1A8L606 V9KDB3 A0A1A8RV89 G3H624 A0A2K6M6H4 W5PKC3
EC Number
2.4.2.-
2.4.2.30
2.4.2.30
Pubmed
19121390
28004739
18362917
19820115
17510324
24845553
+ More
20798317 24508170 30249741 21282665 21719571 21347285 25244985 26823975 20920257 23761445 12364791 14747013 17210077 22992520 25474469 20075255 24495485 25576852 28648823 23537049 28812685 25765539 9215903 26108605 17554307 21551351 15496914 24755649 26483478 15562597 16141072 19468303 15489334 20838655 20010809 19892987 25362486 29704459 24402279 21804562 20809919
20798317 24508170 30249741 21282665 21719571 21347285 25244985 26823975 20920257 23761445 12364791 14747013 17210077 22992520 25474469 20075255 24495485 25576852 28648823 23537049 28812685 25765539 9215903 26108605 17554307 21551351 15496914 24755649 26483478 15562597 16141072 19468303 15489334 20838655 20010809 19892987 25362486 29704459 24402279 21804562 20809919
EMBL
BABH01040453
BABH01040454
BABH01040455
BABH01040456
BABH01040457
BABH01040458
+ More
BABH01040459 BABH01040460 GEZM01035988 JAV83056.1 KQ971311 EEZ98998.2 CH477270 EAT45334.1 NEVH01007830 PNF35046.1 KK852508 KDR22389.1 KK121746 KFM81130.1 GL436716 EFN71576.1 KK107321 QOIP01000002 EZA52680.1 RLU25868.1 GL762889 EFZ20592.1 KQ978060 KYM97722.1 GL888033 EGI69089.1 ADTU01000870 ADTU01000871 KQ983012 KYQ48357.1 KQ978765 KYN28533.1 GBYB01008974 JAG78741.1 KQ976423 KYM88769.1 GFDL01000945 JAV34100.1 UFQT01001759 SSX31685.1 GBRD01005747 JAG60074.1 GBYB01007147 JAG76914.1 GDHC01000890 JAQ17739.1 GGFJ01000930 MBW50071.1 GGFJ01000832 MBW49973.1 AXCM01002785 GGFL01004746 MBW68924.1 KQ434803 KZC05918.1 GGFK01006574 MBW39895.1 GGFL01006400 MBW70578.1 GGFL01004747 MBW68925.1 ADMH02001488 ETN62341.1 GGFL01002620 MBW66798.1 SSX31686.1 AAAB01008987 EAA01120.5 APCN01002058 ACPB03016273 GEDC01012798 JAS24500.1 GFDF01007744 JAV06340.1 JH816438 EKC35064.1 GFDF01007706 JAV06378.1 GBBI01002421 JAC16291.1 HACA01025880 CDW43241.1 AJWK01022423 AJWK01022424 AJWK01022425 GAMC01007023 JAB99532.1 GACK01008571 JAA56463.1 NNAY01000286 OXU29487.1 KZ288271 PBC29847.1 APGK01019502 KB740107 KB631715 ENN81354.1 ERL85556.1 NEDP02004212 OWF46301.1 GANP01007090 JAB77378.1 AFYH01184463 AFYH01184464 AFYH01184465 AFYH01184466 AFYH01184467 AFYH01184468 AFYH01184469 AFYH01184470 AFYH01184471 AFYH01184472 KQ423006 KOF73754.1 JRES01000624 KNC29813.1 HAEB01015887 SBQ62414.1 FR904938 CDQ73931.1 KB108646 ELK28891.1 KE162005 EPQ06383.1 GGMR01017240 MBY29859.1 CYRY02027197 VCW99007.1 JXUM01125409 JXUM01125410 JXUM01125411 KQ566943 KXJ69712.1 AK048860 AC122458 BC057370 HAED01002529 SBQ88374.1 ACTA01108263 ACTA01116263 ACTA01124263 ACTA01132263 ACTA01140263 ACTA01148262 ACTA01156261 ACTA01164260 ACTA01172258 ACTA01180256 JARO02001424 KPP75515.1 HAEG01009311 SBR84186.1 GCES01013909 JAR72414.1 RAZU01000044 RLQ78167.1 GCES01126062 JAQ60260.1 GCES01059208 JAR27115.1 HAEF01001937 SBR39319.1 JW863396 AFO95913.1 HAEH01019937 SBS10000.1 JH000169 EGV98450.1 AMGL01073033 AMGL01073034 AMGL01073035 AMGL01073036 AMGL01073037 AMGL01073038
BABH01040459 BABH01040460 GEZM01035988 JAV83056.1 KQ971311 EEZ98998.2 CH477270 EAT45334.1 NEVH01007830 PNF35046.1 KK852508 KDR22389.1 KK121746 KFM81130.1 GL436716 EFN71576.1 KK107321 QOIP01000002 EZA52680.1 RLU25868.1 GL762889 EFZ20592.1 KQ978060 KYM97722.1 GL888033 EGI69089.1 ADTU01000870 ADTU01000871 KQ983012 KYQ48357.1 KQ978765 KYN28533.1 GBYB01008974 JAG78741.1 KQ976423 KYM88769.1 GFDL01000945 JAV34100.1 UFQT01001759 SSX31685.1 GBRD01005747 JAG60074.1 GBYB01007147 JAG76914.1 GDHC01000890 JAQ17739.1 GGFJ01000930 MBW50071.1 GGFJ01000832 MBW49973.1 AXCM01002785 GGFL01004746 MBW68924.1 KQ434803 KZC05918.1 GGFK01006574 MBW39895.1 GGFL01006400 MBW70578.1 GGFL01004747 MBW68925.1 ADMH02001488 ETN62341.1 GGFL01002620 MBW66798.1 SSX31686.1 AAAB01008987 EAA01120.5 APCN01002058 ACPB03016273 GEDC01012798 JAS24500.1 GFDF01007744 JAV06340.1 JH816438 EKC35064.1 GFDF01007706 JAV06378.1 GBBI01002421 JAC16291.1 HACA01025880 CDW43241.1 AJWK01022423 AJWK01022424 AJWK01022425 GAMC01007023 JAB99532.1 GACK01008571 JAA56463.1 NNAY01000286 OXU29487.1 KZ288271 PBC29847.1 APGK01019502 KB740107 KB631715 ENN81354.1 ERL85556.1 NEDP02004212 OWF46301.1 GANP01007090 JAB77378.1 AFYH01184463 AFYH01184464 AFYH01184465 AFYH01184466 AFYH01184467 AFYH01184468 AFYH01184469 AFYH01184470 AFYH01184471 AFYH01184472 KQ423006 KOF73754.1 JRES01000624 KNC29813.1 HAEB01015887 SBQ62414.1 FR904938 CDQ73931.1 KB108646 ELK28891.1 KE162005 EPQ06383.1 GGMR01017240 MBY29859.1 CYRY02027197 VCW99007.1 JXUM01125409 JXUM01125410 JXUM01125411 KQ566943 KXJ69712.1 AK048860 AC122458 BC057370 HAED01002529 SBQ88374.1 ACTA01108263 ACTA01116263 ACTA01124263 ACTA01132263 ACTA01140263 ACTA01148262 ACTA01156261 ACTA01164260 ACTA01172258 ACTA01180256 JARO02001424 KPP75515.1 HAEG01009311 SBR84186.1 GCES01013909 JAR72414.1 RAZU01000044 RLQ78167.1 GCES01126062 JAQ60260.1 GCES01059208 JAR27115.1 HAEF01001937 SBR39319.1 JW863396 AFO95913.1 HAEH01019937 SBS10000.1 JH000169 EGV98450.1 AMGL01073033 AMGL01073034 AMGL01073035 AMGL01073036 AMGL01073037 AMGL01073038
Proteomes
UP000005204
UP000007266
UP000008820
UP000235965
UP000027135
UP000054359
+ More
UP000000311 UP000053097 UP000279307 UP000078542 UP000007755 UP000005205 UP000075809 UP000078492 UP000078540 UP000076408 UP000075885 UP000075880 UP000075883 UP000076502 UP000075903 UP000000673 UP000007062 UP000075840 UP000015103 UP000005408 UP000002358 UP000092461 UP000215335 UP000242457 UP000019118 UP000030742 UP000005203 UP000242188 UP000008672 UP000053454 UP000037069 UP000075920 UP000001038 UP000095300 UP000005226 UP000007303 UP000193380 UP000075884 UP000078200 UP000075900 UP000069940 UP000249989 UP000076420 UP000261680 UP000000589 UP000001645 UP000008912 UP000002281 UP000069272 UP000034805 UP000233200 UP000265000 UP000273346 UP000252040 UP000248483 UP000001075 UP000233180 UP000002356
UP000000311 UP000053097 UP000279307 UP000078542 UP000007755 UP000005205 UP000075809 UP000078492 UP000078540 UP000076408 UP000075885 UP000075880 UP000075883 UP000076502 UP000075903 UP000000673 UP000007062 UP000075840 UP000015103 UP000005408 UP000002358 UP000092461 UP000215335 UP000242457 UP000019118 UP000030742 UP000005203 UP000242188 UP000008672 UP000053454 UP000037069 UP000075920 UP000001038 UP000095300 UP000005226 UP000007303 UP000193380 UP000075884 UP000078200 UP000075900 UP000069940 UP000249989 UP000076420 UP000261680 UP000000589 UP000001645 UP000008912 UP000002281 UP000069272 UP000034805 UP000233200 UP000265000 UP000273346 UP000252040 UP000248483 UP000001075 UP000233180 UP000002356
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JJ54
A0A1Y1MGQ1
D6WBK7
Q17FK9
A0A2J7R2K4
A0A067RRJ4
+ More
A0A087UUU1 E2A4P2 A0A026W9G4 E9IFR3 A0A151ICA6 F4W9V3 A0A158NZ41 A0A151WKM1 A0A151JP23 A0A0C9QYT9 A0A195BRD1 A0A1Q3G2S5 A0A182YG88 A0A336MPT4 A0A0K8T3I0 A0A182PQV5 A0A0C9R2U6 A0A146ME76 A0A182J3K7 A0A2M4BAR1 A0A2M4BAB1 A0A182MSV2 A0A2M4CUA0 A0A154P3V6 A0A1Y9IS44 A0A2M4AGI6 A0A2M4D0F1 A0A2M4CUJ3 W5JE54 A0A2M4CPG4 A0A336MN10 Q7PYH8 A0A2C9GRX3 T1I1W7 A0A1B6DFS8 A0A1L8DJ06 K1QV17 A0A1L8DIT8 A0A023F473 K7IPE9 A0A0K2UZT5 A0A1B0GJW3 W8BL91 L7LZZ4 A0A232FGN2 A0A2A3EDN9 N6TLM3 A0A088ADB5 A0A210QC45 V5HT76 I6L4N1 A0A0L8GA82 A0A0L0CBM5 A0A1A8FWD1 A0A182VR25 A0A3B3IBB7 A0A1I8PA53 H2TPE9 H2TPF0 H3DNG1 A0A3B3HQ19 A0A3B3I5D9 A0A060X381 L5LS37 A0A182N620 A0A1A9UTL6 S7P8B6 A0A182RXK5 A0A2S2PK85 A0A3P4NNU1 A0A182HBN8 A0A2C9JDF7 A0A384BQD6 Q6PFX9-2 G1N930 A0A1A8HT28 G1LHI1 A0A3Q2I900 A0A182F895 A0A0P7XKI1 A0A2K6NKJ4 A0A1A8PSE2 A0A147A1E8 A0A3L7IJA5 A0A341D1L1 A0A2Y9N107 A0A146QX23 A0A146WDV9 A0A1A8L606 V9KDB3 A0A1A8RV89 G3H624 A0A2K6M6H4 W5PKC3
A0A087UUU1 E2A4P2 A0A026W9G4 E9IFR3 A0A151ICA6 F4W9V3 A0A158NZ41 A0A151WKM1 A0A151JP23 A0A0C9QYT9 A0A195BRD1 A0A1Q3G2S5 A0A182YG88 A0A336MPT4 A0A0K8T3I0 A0A182PQV5 A0A0C9R2U6 A0A146ME76 A0A182J3K7 A0A2M4BAR1 A0A2M4BAB1 A0A182MSV2 A0A2M4CUA0 A0A154P3V6 A0A1Y9IS44 A0A2M4AGI6 A0A2M4D0F1 A0A2M4CUJ3 W5JE54 A0A2M4CPG4 A0A336MN10 Q7PYH8 A0A2C9GRX3 T1I1W7 A0A1B6DFS8 A0A1L8DJ06 K1QV17 A0A1L8DIT8 A0A023F473 K7IPE9 A0A0K2UZT5 A0A1B0GJW3 W8BL91 L7LZZ4 A0A232FGN2 A0A2A3EDN9 N6TLM3 A0A088ADB5 A0A210QC45 V5HT76 I6L4N1 A0A0L8GA82 A0A0L0CBM5 A0A1A8FWD1 A0A182VR25 A0A3B3IBB7 A0A1I8PA53 H2TPE9 H2TPF0 H3DNG1 A0A3B3HQ19 A0A3B3I5D9 A0A060X381 L5LS37 A0A182N620 A0A1A9UTL6 S7P8B6 A0A182RXK5 A0A2S2PK85 A0A3P4NNU1 A0A182HBN8 A0A2C9JDF7 A0A384BQD6 Q6PFX9-2 G1N930 A0A1A8HT28 G1LHI1 A0A3Q2I900 A0A182F895 A0A0P7XKI1 A0A2K6NKJ4 A0A1A8PSE2 A0A147A1E8 A0A3L7IJA5 A0A341D1L1 A0A2Y9N107 A0A146QX23 A0A146WDV9 A0A1A8L606 V9KDB3 A0A1A8RV89 G3H624 A0A2K6M6H4 W5PKC3
PDB
5OWT
E-value=1.38294e-86,
Score=817
Ontologies
GO
GO:0003950
GO:0006471
GO:0090263
GO:0005634
GO:0005737
GO:0070198
GO:1904355
GO:0016055
GO:0051225
GO:0032212
GO:0043565
GO:0006355
GO:0016021
GO:0042593
GO:0031670
GO:0051028
GO:0000209
GO:0005654
GO:0018107
GO:0016604
GO:0005794
GO:0070212
GO:1904357
GO:0018105
GO:1904908
GO:0051301
GO:0008270
GO:0097431
GO:0005643
GO:1990404
GO:0005815
GO:0005829
GO:0042393
GO:0051973
GO:0031965
GO:0000139
GO:1904743
GO:0000781
GO:0000784
GO:0015031
GO:0045944
GO:0070213
GO:0005515
GO:0003677
GO:0006101
GO:0003824
GO:0008152
GO:0016491
GO:0015930
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Golgi apparatus membrane Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Cytoskeleton Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Microtubule organizing center Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Centrosome Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Nuclear pore complex Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Chromosome Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Telomere Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Spindle pole Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Cytoplasm Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Golgi apparatus membrane Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Cytoskeleton Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Microtubule organizing center Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Centrosome Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Nuclear pore complex Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Chromosome Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Telomere Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
Spindle pole Associated with the Golgi and with juxtanuclear SLC2A4/GLUT4-vesicles. A minor proportion is also found at nuclear pore complexes and around the pericentriolar matrix of mitotic centromeres. During interphase, a small fraction of TNKS is found in the nucleus, associated with TERF1. Localizes to spindle poles at mitosis onset via interaction with NUMA1. With evidence from 3 publications.
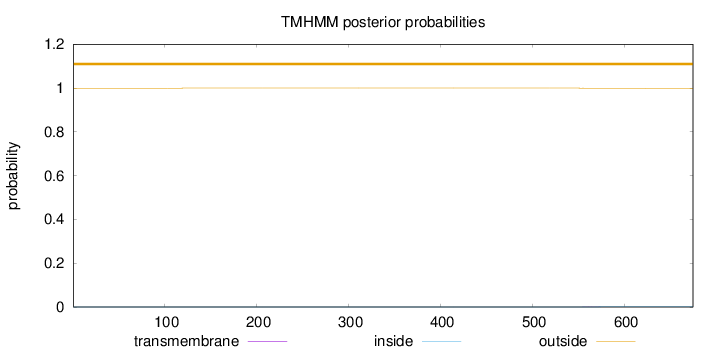
Length:
675
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01161
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.00045
outside
1 - 675
Population Genetic Test Statistics
Pi
19.538582
Theta
19.049921
Tajima's D
-1.813804
CLR
1009.048485
CSRT
0.0264486775661217
Interpretation
Uncertain
