Gene
KWMTBOMO14619 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009551
Annotation
PREDICTED:_serine_proteinase-like_protein_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.836
Sequence
CDS
ATGCGTTCGCTTTTGCTGGCGGTGCTGGTAACAGTCGGTCTCGCCCAGGACACGACCTTAGACCCTGCCTTGCTTCTGAACATCTTCGGAACGCCCCCCACGCCGGCGAAGCCCGGCACGGGGAACCTGGAAGACATCATCGTTAAGCCTACGGAGAGCAACAGCGTGTTTACGGATAAGAACGGGGAATCTTGCAAATGCGTCCCCTACTATTTGTGTAACAAGAACAATGAAGGGGTGGACGTTAATAACGCCAGCGTGACGGGGTGGGGAGTGCTGGATGTCAGGTTCGGCGAAGAAGACTGCCAAGAGAGCGTGGAGATCTGCTGCACTAATCCCATAACGGAACCCGTGCCAAAGCCGCAGCCGGACCCCTCGAAGTTGAAGGGATGCGGCTACAGGAACCCCATGGGGGTCGGAGTGACCATCACCGGAGGGGTGGGTACGGAGGCTCAGTTCGGCGAGTTCCCCTGGGTGGTGGCTCTGCTGGACGCTTTGAACGAATCGTACGCTGGAGTAGGAGTCCTGATACATCCTCAAGTCGTCATGACTGGCGCTCACATCGCTTACAAGTACGCCCCGGGCAATTTGAGGGCGAGAGCCGGAGAATGGGACACCCAGACCATCAAGGAAATGCTGGACCACCAAGTGAGACTCGTCGAGGAAATAATCATACACGAAGATTTCAACACGAAAAGCCTTAAGAACGACGTGGCGCTCCTGCGGATGCACGCGCCCTTCAACCTCGCCGAGCACATCAACATGATCTGCCTCCCGGACCCGGGGGATAGCTTCGACACCAGCAAGAACTGCGTCGCCAACGGCTGGGGGAAGGACGTCTTTGGTCTCCAGGGCAGATACGCGGTGATACTGAAGAAAATCGAAATCGATATGGTGCCGAACCCTCGCTGCAATTCGCTGCTGCAGCGAACCAGACTTGGGACCAGGTTCCGTCTGCACGACAGTTTCGTGTGCGCCGGGGGACAGGAGGGCAGGGACACTTGCCAGGGCGACGGTGGAGCCCCCCTGGCCTGTCCCATAGGTGACAGCCGCTACAAGTTGGCCGGGCTGGTCGCCTGGGGCATCGGGTGCGGGCAGAAGGACGTGCCCGCGGTCTACGCCAACGTGGCCCGCATGAGGTCTTGGGTCGACCGGAAGATGAACGCGTGGGGTTACGGAACCACCACTTACACTATTTAA
Protein
MRSLLLAVLVTVGLAQDTTLDPALLLNIFGTPPTPAKPGTGNLEDIIVKPTESNSVFTDKNGESCKCVPYYLCNKNNEGVDVNNASVTGWGVLDVRFGEEDCQESVEICCTNPITEPVPKPQPDPSKLKGCGYRNPMGVGVTITGGVGTEAQFGEFPWVVALLDALNESYAGVGVLIHPQVVMTGAHIAYKYAPGNLRARAGEWDTQTIKEMLDHQVRLVEEIIIHEDFNTKSLKNDVALLRMHAPFNLAEHINMICLPDPGDSFDTSKNCVANGWGKDVFGLQGRYAVILKKIEIDMVPNPRCNSLLQRTRLGTRFRLHDSFVCAGGQEGRDTCQGDGGAPLACPIGDSRYKLAGLVAWGIGCGQKDVPAVYANVARMRSWVDRKMNAWGYGTTTYTI
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
Q1HPQ5
Q8MUL9
A0A2H1VYD0
A0A2A4K804
A0A194QBB8
I4DPZ1
+ More
A0A2W1BA62 B4KAW8 A0A1J1J6Q9 B4LXF8 B4GM92 A0A0P8XDF8 B5DVP8 A0A1B0CQV9 A0A0A9XG94 A0A1W4VDS9 Q95RS6 B4JRG5 A0A0R1EDK8 A0A0M4EL97 A0A3B0K221 A0A0Q5WPF0 A0A0C9RBZ8 A0A034VVT6 B4G7K9 B4NV29 A0A0J9R5Y2 A0A2M4CY37 A0A0J9R692 A0A1Q3FX14 B4ILN3 A0A154P748 A0A0K8V8L6 A0A0K8VJ89 A0A0Q9X2L0 A0A336KSU8 A0A1I8PTN5 A0A0A1WRZ0 A0A0K8VPE9 A0A1Y9IW08 A0A1J1J5W2 A0A1L8DQQ5 A0A1B6D581 A0A2M4AWK0 A0A1L8DQG3 A0A182L4R5 A0NGL7 A0A182X6T2 Q17HP3 A0A2C9GQL8 A0A182UZX3 B7SVM2 Q9VL01 Q86PE8 A0A2W1BF82 A0A1Y9HE96 A0A0A9Z3K5 A0A0A9VY28 A0A0A9W6K6 A0A3F2YW27 A0A0A9YNY7 A0A3B0K9F1 A0A0K8SQH8 E0VIM5 A0A0K8SMU2 A0A240PMR5 A0A2A4JWR6 A0A182QS83 A0A2M4BPV7 U5ERE0 A0A146LZB7 A0A2P8XJB2 A0A023EU08 A0A1W7R7D2 A0A146LF21 A0A1V1G596 K7J9P4 B3N966 A0A182MK95 A0A182K2D4 U4UIP1 A0A1V1FTC5 N6UK43 A0A2M4CHK8 A0A2J7QQU4 J3JZG5 A0A2S2NKS0 A0A084WHP8 B4NZ90 B4Q8T2 A0A336MWI5 A0A232ET69 B4HWK1 A0A2A3E4S5 V9IJ04 W8BGL7 A0A1B0G1U7 B3MPQ6 A0A385NEX6 A0A067QVT0
A0A2W1BA62 B4KAW8 A0A1J1J6Q9 B4LXF8 B4GM92 A0A0P8XDF8 B5DVP8 A0A1B0CQV9 A0A0A9XG94 A0A1W4VDS9 Q95RS6 B4JRG5 A0A0R1EDK8 A0A0M4EL97 A0A3B0K221 A0A0Q5WPF0 A0A0C9RBZ8 A0A034VVT6 B4G7K9 B4NV29 A0A0J9R5Y2 A0A2M4CY37 A0A0J9R692 A0A1Q3FX14 B4ILN3 A0A154P748 A0A0K8V8L6 A0A0K8VJ89 A0A0Q9X2L0 A0A336KSU8 A0A1I8PTN5 A0A0A1WRZ0 A0A0K8VPE9 A0A1Y9IW08 A0A1J1J5W2 A0A1L8DQQ5 A0A1B6D581 A0A2M4AWK0 A0A1L8DQG3 A0A182L4R5 A0NGL7 A0A182X6T2 Q17HP3 A0A2C9GQL8 A0A182UZX3 B7SVM2 Q9VL01 Q86PE8 A0A2W1BF82 A0A1Y9HE96 A0A0A9Z3K5 A0A0A9VY28 A0A0A9W6K6 A0A3F2YW27 A0A0A9YNY7 A0A3B0K9F1 A0A0K8SQH8 E0VIM5 A0A0K8SMU2 A0A240PMR5 A0A2A4JWR6 A0A182QS83 A0A2M4BPV7 U5ERE0 A0A146LZB7 A0A2P8XJB2 A0A023EU08 A0A1W7R7D2 A0A146LF21 A0A1V1G596 K7J9P4 B3N966 A0A182MK95 A0A182K2D4 U4UIP1 A0A1V1FTC5 N6UK43 A0A2M4CHK8 A0A2J7QQU4 J3JZG5 A0A2S2NKS0 A0A084WHP8 B4NZ90 B4Q8T2 A0A336MWI5 A0A232ET69 B4HWK1 A0A2A3E4S5 V9IJ04 W8BGL7 A0A1B0G1U7 B3MPQ6 A0A385NEX6 A0A067QVT0
Pubmed
19121390
12535678
26354079
22651552
28756777
17994087
+ More
15632085 23185243 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 22936249 25830018 20966253 12364791 14747013 17210077 17510324 19033442 20566863 26823975 29403074 24945155 28410430 20075255 23537049 22516182 24438588 28648823 24495485 18057021 30221452 24845553
15632085 23185243 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 22936249 25830018 20966253 12364791 14747013 17210077 17510324 19033442 20566863 26823975 29403074 24945155 28410430 20075255 23537049 22516182 24438588 28648823 24495485 18057021 30221452 24845553
EMBL
BABH01042089
BABH01042090
DQ443347
ABF51436.1
AF518768
AAM69353.1
+ More
ODYU01005195 SOQ45813.1 NWSH01000060 PCG80068.1 KQ459249 KPJ02285.1 AK403853 BAM19981.1 KZ150189 PZC72429.1 CH933806 EDW14646.1 CVRI01000070 CRL07166.1 CH940650 EDW67836.2 CH479185 EDW37966.1 CH902623 KPU72696.1 CM000070 EDY68456.1 KRT01050.1 AJWK01024105 AJWK01024106 AJWK01024107 GBHO01024948 JAG18656.1 AY061167 BT022896 AE014296 AAL28715.1 AAY55312.1 EAA46220.1 CH916373 EDV94355.1 CH892676 KRK05537.1 CP012526 ALC46027.1 OUUW01000014 SPP88345.1 CH954177 KQS71087.1 GBYB01010652 JAG80419.1 GAKP01012987 GAKP01012985 JAC45965.1 CH479180 EDW28409.1 CH985579 CM002911 EDX15901.1 KMY91511.1 KMY91513.1 KMY91515.1 GGFL01006066 MBW70244.1 KMY91516.1 GFDL01002926 JAV32119.1 CH480881 EDW54724.1 KQ434829 KZC07692.1 GDHF01017181 JAI35133.1 GDHF01013360 JAI38954.1 CH964272 KRG00138.1 UFQS01000960 UFQT01000960 UFQT01003904 SSX07970.1 SSX28204.1 SSX35355.1 GBXI01013017 GBXI01012605 GBXI01000118 JAD01275.1 JAD01687.1 JAD14174.1 GDHF01011849 JAI40465.1 CRL07164.1 GFDF01005367 JAV08717.1 GEDC01016436 JAS20862.1 GGFK01011761 MBW45082.1 GFDF01005366 JAV08718.1 AAAB01008986 EAL38870.1 CH477246 EAT46196.1 EAT46197.1 APCN01002395 EU770391 ACI32835.1 AE014134 BT044284 AAF52904.1 ACH92349.1 BT003168 AAO24923.1 KZ150319 PZC71430.1 GBHO01044513 GBHO01038574 GBHO01007209 GBHO01007204 JAF99090.1 JAG05030.1 JAG36395.1 JAG36400.1 GBHO01044521 GBHO01038579 GBHO01038553 GBHO01022104 JAF99082.1 JAG05025.1 JAG05051.1 JAG21500.1 GBHO01044520 GBHO01040165 GBHO01008846 GBHO01007201 JAF99083.1 JAG03439.1 JAG34758.1 JAG36403.1 GBHO01044518 GBHO01008852 GBHO01008851 GBHO01008848 JAF99085.1 JAG34752.1 JAG34753.1 JAG34756.1 OUUW01000006 SPP81651.1 GBRD01010286 GBRD01010284 JAG55538.1 DS235201 EEB13231.1 GBRD01011162 GBRD01010285 JAG54662.1 NWSH01000512 PCG75933.1 AXCN02001108 GGFJ01005667 MBW54808.1 GANO01003790 JAB56081.1 GDHC01005736 JAQ12893.1 PYGN01001938 PSN32091.1 GAPW01001674 JAC11924.1 GEHC01000587 JAV47058.1 GDHC01013032 JAQ05597.1 FX985353 BAX07366.1 AAZX01008083 EDV58501.1 AXCM01000676 KB632375 ERL93884.1 FX985352 BAX07365.1 APGK01030042 APGK01030043 APGK01030044 APGK01030045 KB740754 ENN79052.1 GGFL01000652 MBW64830.1 NEVH01012082 PNF30937.1 BT128647 AEE63604.1 GGMR01005162 MBY17781.1 ATLV01023866 KE525347 KFB49742.1 CM000157 EDW88785.1 CM000361 CM002910 EDX04497.1 KMY89480.1 UFQT01003062 SSX34500.1 NNAY01002322 OXU21553.1 CH480818 EDW52396.1 KZ288370 PBC26753.1 JR049796 AEY61083.1 GAMC01017753 GAMC01017751 JAB88804.1 CCAG010021201 CH902620 EDV32304.2 KPU73895.1 MH261257 AYA71582.1 KK853313 KDR08623.1
ODYU01005195 SOQ45813.1 NWSH01000060 PCG80068.1 KQ459249 KPJ02285.1 AK403853 BAM19981.1 KZ150189 PZC72429.1 CH933806 EDW14646.1 CVRI01000070 CRL07166.1 CH940650 EDW67836.2 CH479185 EDW37966.1 CH902623 KPU72696.1 CM000070 EDY68456.1 KRT01050.1 AJWK01024105 AJWK01024106 AJWK01024107 GBHO01024948 JAG18656.1 AY061167 BT022896 AE014296 AAL28715.1 AAY55312.1 EAA46220.1 CH916373 EDV94355.1 CH892676 KRK05537.1 CP012526 ALC46027.1 OUUW01000014 SPP88345.1 CH954177 KQS71087.1 GBYB01010652 JAG80419.1 GAKP01012987 GAKP01012985 JAC45965.1 CH479180 EDW28409.1 CH985579 CM002911 EDX15901.1 KMY91511.1 KMY91513.1 KMY91515.1 GGFL01006066 MBW70244.1 KMY91516.1 GFDL01002926 JAV32119.1 CH480881 EDW54724.1 KQ434829 KZC07692.1 GDHF01017181 JAI35133.1 GDHF01013360 JAI38954.1 CH964272 KRG00138.1 UFQS01000960 UFQT01000960 UFQT01003904 SSX07970.1 SSX28204.1 SSX35355.1 GBXI01013017 GBXI01012605 GBXI01000118 JAD01275.1 JAD01687.1 JAD14174.1 GDHF01011849 JAI40465.1 CRL07164.1 GFDF01005367 JAV08717.1 GEDC01016436 JAS20862.1 GGFK01011761 MBW45082.1 GFDF01005366 JAV08718.1 AAAB01008986 EAL38870.1 CH477246 EAT46196.1 EAT46197.1 APCN01002395 EU770391 ACI32835.1 AE014134 BT044284 AAF52904.1 ACH92349.1 BT003168 AAO24923.1 KZ150319 PZC71430.1 GBHO01044513 GBHO01038574 GBHO01007209 GBHO01007204 JAF99090.1 JAG05030.1 JAG36395.1 JAG36400.1 GBHO01044521 GBHO01038579 GBHO01038553 GBHO01022104 JAF99082.1 JAG05025.1 JAG05051.1 JAG21500.1 GBHO01044520 GBHO01040165 GBHO01008846 GBHO01007201 JAF99083.1 JAG03439.1 JAG34758.1 JAG36403.1 GBHO01044518 GBHO01008852 GBHO01008851 GBHO01008848 JAF99085.1 JAG34752.1 JAG34753.1 JAG34756.1 OUUW01000006 SPP81651.1 GBRD01010286 GBRD01010284 JAG55538.1 DS235201 EEB13231.1 GBRD01011162 GBRD01010285 JAG54662.1 NWSH01000512 PCG75933.1 AXCN02001108 GGFJ01005667 MBW54808.1 GANO01003790 JAB56081.1 GDHC01005736 JAQ12893.1 PYGN01001938 PSN32091.1 GAPW01001674 JAC11924.1 GEHC01000587 JAV47058.1 GDHC01013032 JAQ05597.1 FX985353 BAX07366.1 AAZX01008083 EDV58501.1 AXCM01000676 KB632375 ERL93884.1 FX985352 BAX07365.1 APGK01030042 APGK01030043 APGK01030044 APGK01030045 KB740754 ENN79052.1 GGFL01000652 MBW64830.1 NEVH01012082 PNF30937.1 BT128647 AEE63604.1 GGMR01005162 MBY17781.1 ATLV01023866 KE525347 KFB49742.1 CM000157 EDW88785.1 CM000361 CM002910 EDX04497.1 KMY89480.1 UFQT01003062 SSX34500.1 NNAY01002322 OXU21553.1 CH480818 EDW52396.1 KZ288370 PBC26753.1 JR049796 AEY61083.1 GAMC01017753 GAMC01017751 JAB88804.1 CCAG010021201 CH902620 EDV32304.2 KPU73895.1 MH261257 AYA71582.1 KK853313 KDR08623.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000009192
UP000183832
UP000008792
+ More
UP000008744 UP000007801 UP000001819 UP000092461 UP000192221 UP000000803 UP000001070 UP000002282 UP000092553 UP000268350 UP000008711 UP000000304 UP000001292 UP000076502 UP000007798 UP000095300 UP000075920 UP000075882 UP000007062 UP000076407 UP000008820 UP000075840 UP000075903 UP000075900 UP000075884 UP000009046 UP000075880 UP000075886 UP000245037 UP000002358 UP000075883 UP000075881 UP000030742 UP000019118 UP000235965 UP000030765 UP000215335 UP000242457 UP000092444 UP000027135
UP000008744 UP000007801 UP000001819 UP000092461 UP000192221 UP000000803 UP000001070 UP000002282 UP000092553 UP000268350 UP000008711 UP000000304 UP000001292 UP000076502 UP000007798 UP000095300 UP000075920 UP000075882 UP000007062 UP000076407 UP000008820 UP000075840 UP000075903 UP000075900 UP000075884 UP000009046 UP000075880 UP000075886 UP000245037 UP000002358 UP000075883 UP000075881 UP000030742 UP000019118 UP000235965 UP000030765 UP000215335 UP000242457 UP000092444 UP000027135
Interpro
Gene 3D
CDD
ProteinModelPortal
Q1HPQ5
Q8MUL9
A0A2H1VYD0
A0A2A4K804
A0A194QBB8
I4DPZ1
+ More
A0A2W1BA62 B4KAW8 A0A1J1J6Q9 B4LXF8 B4GM92 A0A0P8XDF8 B5DVP8 A0A1B0CQV9 A0A0A9XG94 A0A1W4VDS9 Q95RS6 B4JRG5 A0A0R1EDK8 A0A0M4EL97 A0A3B0K221 A0A0Q5WPF0 A0A0C9RBZ8 A0A034VVT6 B4G7K9 B4NV29 A0A0J9R5Y2 A0A2M4CY37 A0A0J9R692 A0A1Q3FX14 B4ILN3 A0A154P748 A0A0K8V8L6 A0A0K8VJ89 A0A0Q9X2L0 A0A336KSU8 A0A1I8PTN5 A0A0A1WRZ0 A0A0K8VPE9 A0A1Y9IW08 A0A1J1J5W2 A0A1L8DQQ5 A0A1B6D581 A0A2M4AWK0 A0A1L8DQG3 A0A182L4R5 A0NGL7 A0A182X6T2 Q17HP3 A0A2C9GQL8 A0A182UZX3 B7SVM2 Q9VL01 Q86PE8 A0A2W1BF82 A0A1Y9HE96 A0A0A9Z3K5 A0A0A9VY28 A0A0A9W6K6 A0A3F2YW27 A0A0A9YNY7 A0A3B0K9F1 A0A0K8SQH8 E0VIM5 A0A0K8SMU2 A0A240PMR5 A0A2A4JWR6 A0A182QS83 A0A2M4BPV7 U5ERE0 A0A146LZB7 A0A2P8XJB2 A0A023EU08 A0A1W7R7D2 A0A146LF21 A0A1V1G596 K7J9P4 B3N966 A0A182MK95 A0A182K2D4 U4UIP1 A0A1V1FTC5 N6UK43 A0A2M4CHK8 A0A2J7QQU4 J3JZG5 A0A2S2NKS0 A0A084WHP8 B4NZ90 B4Q8T2 A0A336MWI5 A0A232ET69 B4HWK1 A0A2A3E4S5 V9IJ04 W8BGL7 A0A1B0G1U7 B3MPQ6 A0A385NEX6 A0A067QVT0
A0A2W1BA62 B4KAW8 A0A1J1J6Q9 B4LXF8 B4GM92 A0A0P8XDF8 B5DVP8 A0A1B0CQV9 A0A0A9XG94 A0A1W4VDS9 Q95RS6 B4JRG5 A0A0R1EDK8 A0A0M4EL97 A0A3B0K221 A0A0Q5WPF0 A0A0C9RBZ8 A0A034VVT6 B4G7K9 B4NV29 A0A0J9R5Y2 A0A2M4CY37 A0A0J9R692 A0A1Q3FX14 B4ILN3 A0A154P748 A0A0K8V8L6 A0A0K8VJ89 A0A0Q9X2L0 A0A336KSU8 A0A1I8PTN5 A0A0A1WRZ0 A0A0K8VPE9 A0A1Y9IW08 A0A1J1J5W2 A0A1L8DQQ5 A0A1B6D581 A0A2M4AWK0 A0A1L8DQG3 A0A182L4R5 A0NGL7 A0A182X6T2 Q17HP3 A0A2C9GQL8 A0A182UZX3 B7SVM2 Q9VL01 Q86PE8 A0A2W1BF82 A0A1Y9HE96 A0A0A9Z3K5 A0A0A9VY28 A0A0A9W6K6 A0A3F2YW27 A0A0A9YNY7 A0A3B0K9F1 A0A0K8SQH8 E0VIM5 A0A0K8SMU2 A0A240PMR5 A0A2A4JWR6 A0A182QS83 A0A2M4BPV7 U5ERE0 A0A146LZB7 A0A2P8XJB2 A0A023EU08 A0A1W7R7D2 A0A146LF21 A0A1V1G596 K7J9P4 B3N966 A0A182MK95 A0A182K2D4 U4UIP1 A0A1V1FTC5 N6UK43 A0A2M4CHK8 A0A2J7QQU4 J3JZG5 A0A2S2NKS0 A0A084WHP8 B4NZ90 B4Q8T2 A0A336MWI5 A0A232ET69 B4HWK1 A0A2A3E4S5 V9IJ04 W8BGL7 A0A1B0G1U7 B3MPQ6 A0A385NEX6 A0A067QVT0
PDB
2B9L
E-value=1.71268e-69,
Score=667
Ontologies
GO
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.998040
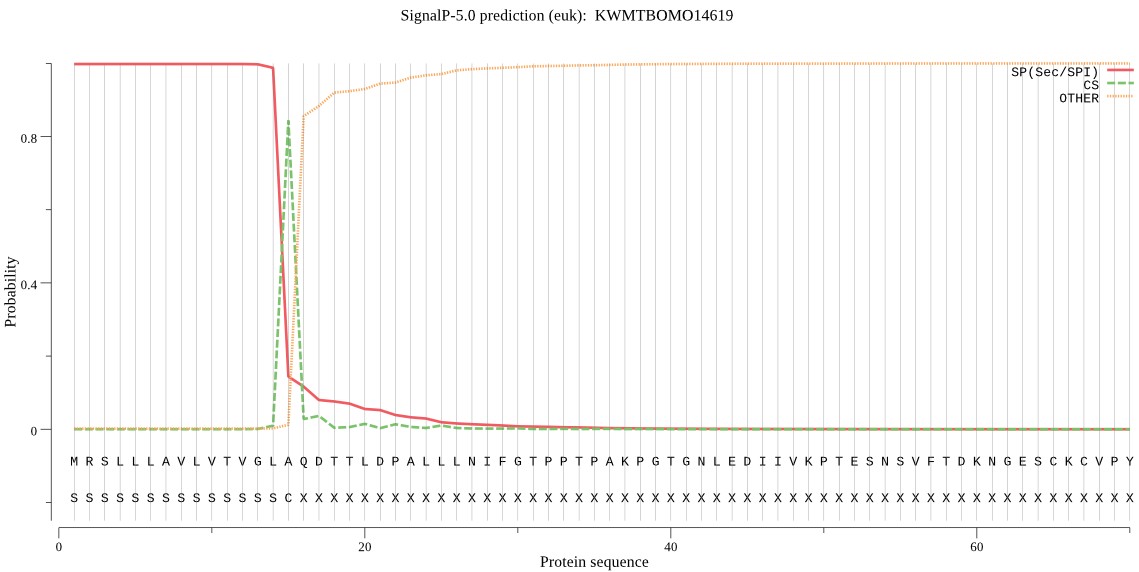
Length:
399
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.22066
Exp number, first 60 AAs:
0.11596
Total prob of N-in:
0.00744
outside
1 - 399
Population Genetic Test Statistics
Pi
253.239116
Theta
152.878138
Tajima's D
1.846381
CLR
0.119188
CSRT
0.858657067146643
Interpretation
Uncertain
