Gene
KWMTBOMO14616 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008776
Annotation
PREDICTED:_metal_transporter_CNNM4-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.082 Nuclear Reliability : 1.331 PlasmaMembrane Reliability : 1.238
Sequence
CDS
ATGGTAGGGCGGAACAACCCCTTTGGCGGCCAGTGGGTGCATCAAGGCTCCGAATATTATTTGCAGCCGAAAAACGATTCTCTTTCAGAGGAGAATAATCGATCGCAAAAGCCGAGGTCTCTCAACGAACTCCAGGAAAACGAAGATAACTGGATCGACCTCTCCCACGACATCGACGTGAATTATCTTCCCAGAGCCAGACCTGCTCGTAATGATAATCCAGATGTAGAAATATTTAGACACGATGATTATATAGCTCTGGATGGAACCGATAAAGTTTGGGATGAAAAGAATAATTTGGGACGGTTTAAAAGAGCTGATGGGATTGCGGCTAAAGCTGCGGAGAGAAGCTCGACAGATTTAGACAATGTCGTGTTAAACAAAAACGCTTCTAATGATGATGGCAGTAAAGATGACACTCGAACAAGAGCTCAAGGATCTGCGAGTGAATTGTTCTCAGAGGAATTGATAAGAAAATCTCGTCAAATAGTAGATTCGAATGATTTTACGAGTATGCGAGCGGCCCCTATCGATATTTTCATCGAAGGAATCAGGATCGAAGAGTCCGAGAAAGAACCACATATTGAAGACGGAATACCGAGTGTGCTGGCCGATACTGCAGTTACGATCAGGGTATTCGGCGATGGGTTAACTTCGAAAACTGCCATCGCTTTCACACGACAGGACCTGCCGTACGGCCAACCCTGCAAGTTTCTACTCATGGGAGAGTATATGGTAAAAGAAGGCAGCGTCACCACCAGATCCGCGTTGTTCGATGTAACAGCACCCCCCTCACAGGAGGGCTCTAAGATCTACATCTGCGCTAAGAATCTCCAGGCAGGGGCTGATCACCCGACCAAGGACAAGGAAAATTACTATCACCAGGGAACTGAATCCTGGAAGGCGTTTGCTACCCACAACACCCTGCTGCCGCTCTGGGTGTCGCTGAGTTTGATTCTGATATGCCTGACATTCTCTGCGCTTTTCTCTGGGTTGAATCTCGGTCTGATGTCATTGGACAGGACCGAACTGAAGATCATAGCCAATACCGGTACGGAAACGGAGAGGAAATACGCCAGAGCCATCATGCCCGTCCGGGATCATGGAAATTACTTGCTGTGCAGCATATTGTTGGGGAATGTGGCTGTGAATTCGACTTTCACGATTTTGTTGGATGAACTGACTTCGGGCCTCTTTGCTGTTATATTCTCGACCCTGGCTATCGTACTCCTGGGCGAGATAACTCCCCAAGCCATATGTTCGAGACACGGTCTTATGATTGGTGCCAAGAGTATCATGATTACGAAGGCGGTTATGGCCCTGACGGCGCCGATGGCGTATCCTGTGAGTAAACTCCTGGACTATTTCTTAGGCGAGGAGATAGGCAGCGTTTACAACAGGGAACGTCTAAAGGAGCTTATCAAGGTGACAACTGGGGACTTGGACAAGGACGAGGTGAACATCATATCCGGCGCCCTCGAGATGAGGAAGAAGACCGTGTCAGACGTCATGACGAAGATGGAGGACGTGTTCATGTTGCCGATAACAAGCGTCCTGGACTTCGAGACGATGTCGGAAATTGTAAAGTCGGGCTACTCCCGCATCCCGGTGTACGAGGGTCACCGCGGGAACATCGTGACGGTGCTCTTCATCAAGGACCTCGCCTTCGTCGATCCCGACGACAACACGCCCCTTAGGACCCTCTGCCAGTACTACCAGAACCCTTGCAACTTCGTCTTCGAAGACGTCACCTTGGATGTCATGTTCAAACAGTTCAAAGAAGGTCACAAAGGACACATGGCGTTCGTCCATCGCATAAACAACGAGGGCGAGGGAGACCCCTTCTACGAGACCATAGGGCTGGTGACCCTGGAAGATGTCATCGAAGAGATGATACAAGCAGAAATCGTCGATGAGACTGATGTGTTCATGGACAACCGCAGCAAGCGACGCAGGAACCGCCCCTCGCACAAGCTCCAGGACTTCGCCGCGTTCGCGGAGAGGCACGAGAACCAGAGGATCCACATCTCGCCCCAGCTCACCCTGGCCACGTTCCAGTTCCTGAGCACGAGCGTAGACGCGTTCCGTCCGGACGTAGTGTCGGAGACGGTTCTGAGACGTCTCCTGAGGCAAGACGTCATCCAGCACATCAAGCTTAAGGGGAAGACCAGACGGGATTCGGCCACATACGTTTACCATCAGGGGAAACCGGTCGATTACTTCGTTTTAATCTTAGAAGGACGAGTCGAAGTTACCGTAGGACGGGAGAACTTGATCTTTGAAGCTGGACCCTTCACATACTTCGGTGTACAAGCCCTTACACAGAACGTTGGAGTTGCCGAGTCTCCTACACCATCAGCGATGGGTTCCCTACAGAACATCAATATGGACTCGATGTTGCGACACACTTTTGTACCGGACTATTCGGTTCGCGCTATCTCTTCCGAACTTTTCTATCTCACTGTGAAGCGCGCTCTGTACCTGGCTGCGAAAAGGGCGACCCTAATGGAGAAGGGCGCCCTCTCCAAGGGCACGACCAACGAACAATTCGACACTGAAGTCGACAAGTTACTCCAGTCTGTAGACGAGGACATCAGCGTGAGCGGGGAGCACAGGACTCCATCGAGACAGGTATCGCCCAATCCTCCACCAGTGTCGGCTTCGCCCCACGTTCGGGCAACTTTCGCGTCGCCCGACAACAATGGCGACGTTTACACGACCCACAACCTGAAGGCCGACGAGGAGGAGAAACTTCTGAAGTGA
Protein
MVGRNNPFGGQWVHQGSEYYLQPKNDSLSEENNRSQKPRSLNELQENEDNWIDLSHDIDVNYLPRARPARNDNPDVEIFRHDDYIALDGTDKVWDEKNNLGRFKRADGIAAKAAERSSTDLDNVVLNKNASNDDGSKDDTRTRAQGSASELFSEELIRKSRQIVDSNDFTSMRAAPIDIFIEGIRIEESEKEPHIEDGIPSVLADTAVTIRVFGDGLTSKTAIAFTRQDLPYGQPCKFLLMGEYMVKEGSVTTRSALFDVTAPPSQEGSKIYICAKNLQAGADHPTKDKENYYHQGTESWKAFATHNTLLPLWVSLSLILICLTFSALFSGLNLGLMSLDRTELKIIANTGTETERKYARAIMPVRDHGNYLLCSILLGNVAVNSTFTILLDELTSGLFAVIFSTLAIVLLGEITPQAICSRHGLMIGAKSIMITKAVMALTAPMAYPVSKLLDYFLGEEIGSVYNRERLKELIKVTTGDLDKDEVNIISGALEMRKKTVSDVMTKMEDVFMLPITSVLDFETMSEIVKSGYSRIPVYEGHRGNIVTVLFIKDLAFVDPDDNTPLRTLCQYYQNPCNFVFEDVTLDVMFKQFKEGHKGHMAFVHRINNEGEGDPFYETIGLVTLEDVIEEMIQAEIVDETDVFMDNRSKRRRNRPSHKLQDFAAFAERHENQRIHISPQLTLATFQFLSTSVDAFRPDVVSETVLRRLLRQDVIQHIKLKGKTRRDSATYVYHQGKPVDYFVLILEGRVEVTVGRENLIFEAGPFTYFGVQALTQNVGVAESPTPSAMGSLQNINMDSMLRHTFVPDYSVRAISSELFYLTVKRALYLAAKRATLMEKGALSKGTTNEQFDTEVDKLLQSVDEDISVSGEHRTPSRQVSPNPPPVSASPHVRATFASPDNNGDVYTTHNLKADEEEKLLK
Summary
Uniprot
A0A2A4K7S3
A0A194QA38
A0A212F849
A0A3S2TFD0
H9JJ62
A0A2A4JS31
+ More
A0A2H1VKG4 A0A088AEY8 A0A0N0BJY2 A0A0L7QKM1 A0A2A3EE16 A0A310S7G1 A0A154PFC6 A0A1L8DDT5 A0A182GUV4 Q16HD8 A0A195C5G0 A0A2J7RPU8 A0A023EVD1 E2BD06 A0A195EXQ7 A0A3L8E0X0 E9J5L7 A0A151J4V4 A0A026WNK7 A0A151WQ63 F4WTD0 A0A195BB62 A0A158NVU0 A0A1Q3G0I8 D6W9C1 A0A1Q3G0K9 E2AJ07 A0A067QMK4 K7J1W6 A0A1I8NKL3 A0A1I8MXE5 A0A1I8PUF5 A0A1I8PUG6 T1PG40 A0A1B6H3Z8 A0A1B6G206 A0A1Y1JZ23 A0A3B0JTY0 A0A1W4XF17 A0A1B6FH96 A0A1B6F7C0 A0A0Q9WAQ1 A0A232FCI9 A0A2H8TPI9 A0A0L0BQR4 A0A0P8XKN0 W8B844 A0A2S2NWZ3 A0A1Y1JR54 J9JL81 A0A1Y1JU74 U4U987 A0A0Q5WLR7 A0A0B7P9G0 N6TAW9 A0A0M4EQT3 A0A034W374 A0A0K8W7G5 A0A0R1ED38 A0A0R1EAX5 A0A0R1E7R6 A0A0Q9X6C8 A0A1J1IK57 A0A1A9XGV8 A0A0A1WZ12 A0A1B6DP17 A0A2M4BBT6 A0A2M4BBM2 W5JKZ4 A0A2M3Z3Q6 A0A2M4BC05 A0A2M3ZGL8 A0A1B0A504 A0A2M4ABE8 A0A1B0GLB6 A0A2M4AB51 A0A2M4ABR6 A0A1B6DR42 A0A182FRV8 T1I5R7 A0A1B6KU35 A0A2M3Z561 E0VYD8 A0A2R7X6X1 B4JAK1 A0A146KNK0 A0A084WSG8 A0A0A9WHS3 A0A146LPK8 A0A182NFP3
A0A2H1VKG4 A0A088AEY8 A0A0N0BJY2 A0A0L7QKM1 A0A2A3EE16 A0A310S7G1 A0A154PFC6 A0A1L8DDT5 A0A182GUV4 Q16HD8 A0A195C5G0 A0A2J7RPU8 A0A023EVD1 E2BD06 A0A195EXQ7 A0A3L8E0X0 E9J5L7 A0A151J4V4 A0A026WNK7 A0A151WQ63 F4WTD0 A0A195BB62 A0A158NVU0 A0A1Q3G0I8 D6W9C1 A0A1Q3G0K9 E2AJ07 A0A067QMK4 K7J1W6 A0A1I8NKL3 A0A1I8MXE5 A0A1I8PUF5 A0A1I8PUG6 T1PG40 A0A1B6H3Z8 A0A1B6G206 A0A1Y1JZ23 A0A3B0JTY0 A0A1W4XF17 A0A1B6FH96 A0A1B6F7C0 A0A0Q9WAQ1 A0A232FCI9 A0A2H8TPI9 A0A0L0BQR4 A0A0P8XKN0 W8B844 A0A2S2NWZ3 A0A1Y1JR54 J9JL81 A0A1Y1JU74 U4U987 A0A0Q5WLR7 A0A0B7P9G0 N6TAW9 A0A0M4EQT3 A0A034W374 A0A0K8W7G5 A0A0R1ED38 A0A0R1EAX5 A0A0R1E7R6 A0A0Q9X6C8 A0A1J1IK57 A0A1A9XGV8 A0A0A1WZ12 A0A1B6DP17 A0A2M4BBT6 A0A2M4BBM2 W5JKZ4 A0A2M3Z3Q6 A0A2M4BC05 A0A2M3ZGL8 A0A1B0A504 A0A2M4ABE8 A0A1B0GLB6 A0A2M4AB51 A0A2M4ABR6 A0A1B6DR42 A0A182FRV8 T1I5R7 A0A1B6KU35 A0A2M3Z561 E0VYD8 A0A2R7X6X1 B4JAK1 A0A146KNK0 A0A084WSG8 A0A0A9WHS3 A0A146LPK8 A0A182NFP3
Pubmed
26354079
22118469
19121390
26483478
17510324
24945155
+ More
20798317 30249741 21282665 24508170 21719571 21347285 18362917 19820115 24845553 20075255 25315136 28004739 17994087 28648823 26108605 24495485 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 17550304 25830018 20920257 23761445 20566863 26823975 24438588 25401762
20798317 30249741 21282665 24508170 21719571 21347285 18362917 19820115 24845553 20075255 25315136 28004739 17994087 28648823 26108605 24495485 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 17550304 25830018 20920257 23761445 20566863 26823975 24438588 25401762
EMBL
NWSH01000060
PCG80066.1
KQ459249
KPJ02289.1
AGBW02009783
OWR49924.1
+ More
RSAL01000199 RVE44514.1 BABH01040476 BABH01040477 NWSH01000720 PCG74629.1 ODYU01003035 SOQ41307.1 KQ435710 KOX79812.1 KQ414940 KOC59188.1 KZ288287 PBC29416.1 KQ777939 OAD52039.1 KQ434892 KZC10512.1 GFDF01009584 JAV04500.1 JXUM01089765 KQ563796 KXJ73288.1 CH478182 EAT33657.1 KQ978231 KYM96089.1 NEVH01001355 PNF42861.1 GAPW01000183 JAC13415.1 GL447495 EFN86431.1 KQ981940 KYN32677.1 QOIP01000001 RLU26364.1 GL768163 EFZ11891.1 KQ980079 KYN17809.1 KK107159 EZA56684.1 KQ982850 KYQ49943.1 GL888334 EGI62522.1 KQ976529 KYM81791.1 ADTU01027491 ADTU01027492 GFDL01001745 JAV33300.1 KQ971312 EEZ98477.1 GFDL01001739 JAV33306.1 GL439921 EFN66596.1 KK853149 KDR10673.1 KA647782 AFP62411.1 GECZ01000402 JAS69367.1 GECZ01013304 JAS56465.1 GEZM01102946 JAV51687.1 OUUW01000010 SPP85574.1 GECZ01020191 JAS49578.1 GECZ01023653 JAS46116.1 CH940649 KRF81674.1 NNAY01000444 OXU28342.1 GFXV01004124 MBW15929.1 JRES01001511 KNC22356.1 CH902628 KPU75331.1 GAMC01017159 JAB89396.1 GGMR01008607 MBY21226.1 GEZM01102944 JAV51689.1 ABLF02030588 ABLF02030600 ABLF02030602 ABLF02030604 GEZM01102945 JAV51688.1 KB631924 ERL87181.1 CH954177 KQS71083.1 AE013599 AIG63332.1 EDP28148.2 EDP28149.2 ELP57407.1 APGK01044408 APGK01044409 APGK01044410 APGK01044411 KB741026 ENN74893.1 CP012523 ALC39157.1 GAKP01008896 JAC50056.1 GDHF01005181 JAI47133.1 CH891640 KRK05076.1 KRK05075.1 KRK05077.1 CH933807 KRG03803.1 CVRI01000048 CRK98833.1 GBXI01010406 JAD03886.1 GEDC01009880 JAS27418.1 GGFJ01001300 MBW50441.1 GGFJ01001304 MBW50445.1 ADMH02001227 ETN63585.1 GGFM01002384 MBW23135.1 GGFJ01001207 MBW50348.1 GGFM01006966 MBW27717.1 GGFK01004617 MBW37938.1 AJWK01034728 AJWK01034729 AJWK01034730 AJWK01034731 GGFK01004694 MBW38015.1 GGFK01004914 MBW38235.1 GEDC01009151 JAS28147.1 ACPB03006135 GEBQ01025213 JAT14764.1 GGFM01002893 MBW23644.1 DS235845 EEB18394.1 KK857470 PTY27200.1 CH916368 EDW02787.1 GDHC01022009 JAP96619.1 ATLV01026534 KE525415 KFB53162.1 GBHO01039189 JAG04415.1 GDHC01009380 JAQ09249.1
RSAL01000199 RVE44514.1 BABH01040476 BABH01040477 NWSH01000720 PCG74629.1 ODYU01003035 SOQ41307.1 KQ435710 KOX79812.1 KQ414940 KOC59188.1 KZ288287 PBC29416.1 KQ777939 OAD52039.1 KQ434892 KZC10512.1 GFDF01009584 JAV04500.1 JXUM01089765 KQ563796 KXJ73288.1 CH478182 EAT33657.1 KQ978231 KYM96089.1 NEVH01001355 PNF42861.1 GAPW01000183 JAC13415.1 GL447495 EFN86431.1 KQ981940 KYN32677.1 QOIP01000001 RLU26364.1 GL768163 EFZ11891.1 KQ980079 KYN17809.1 KK107159 EZA56684.1 KQ982850 KYQ49943.1 GL888334 EGI62522.1 KQ976529 KYM81791.1 ADTU01027491 ADTU01027492 GFDL01001745 JAV33300.1 KQ971312 EEZ98477.1 GFDL01001739 JAV33306.1 GL439921 EFN66596.1 KK853149 KDR10673.1 KA647782 AFP62411.1 GECZ01000402 JAS69367.1 GECZ01013304 JAS56465.1 GEZM01102946 JAV51687.1 OUUW01000010 SPP85574.1 GECZ01020191 JAS49578.1 GECZ01023653 JAS46116.1 CH940649 KRF81674.1 NNAY01000444 OXU28342.1 GFXV01004124 MBW15929.1 JRES01001511 KNC22356.1 CH902628 KPU75331.1 GAMC01017159 JAB89396.1 GGMR01008607 MBY21226.1 GEZM01102944 JAV51689.1 ABLF02030588 ABLF02030600 ABLF02030602 ABLF02030604 GEZM01102945 JAV51688.1 KB631924 ERL87181.1 CH954177 KQS71083.1 AE013599 AIG63332.1 EDP28148.2 EDP28149.2 ELP57407.1 APGK01044408 APGK01044409 APGK01044410 APGK01044411 KB741026 ENN74893.1 CP012523 ALC39157.1 GAKP01008896 JAC50056.1 GDHF01005181 JAI47133.1 CH891640 KRK05076.1 KRK05075.1 KRK05077.1 CH933807 KRG03803.1 CVRI01000048 CRK98833.1 GBXI01010406 JAD03886.1 GEDC01009880 JAS27418.1 GGFJ01001300 MBW50441.1 GGFJ01001304 MBW50445.1 ADMH02001227 ETN63585.1 GGFM01002384 MBW23135.1 GGFJ01001207 MBW50348.1 GGFM01006966 MBW27717.1 GGFK01004617 MBW37938.1 AJWK01034728 AJWK01034729 AJWK01034730 AJWK01034731 GGFK01004694 MBW38015.1 GGFK01004914 MBW38235.1 GEDC01009151 JAS28147.1 ACPB03006135 GEBQ01025213 JAT14764.1 GGFM01002893 MBW23644.1 DS235845 EEB18394.1 KK857470 PTY27200.1 CH916368 EDW02787.1 GDHC01022009 JAP96619.1 ATLV01026534 KE525415 KFB53162.1 GBHO01039189 JAG04415.1 GDHC01009380 JAQ09249.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000283053
UP000005204
UP000005203
+ More
UP000053105 UP000053825 UP000242457 UP000076502 UP000069940 UP000249989 UP000008820 UP000078542 UP000235965 UP000008237 UP000078541 UP000279307 UP000078492 UP000053097 UP000075809 UP000007755 UP000078540 UP000005205 UP000007266 UP000000311 UP000027135 UP000002358 UP000095301 UP000095300 UP000268350 UP000192223 UP000008792 UP000215335 UP000037069 UP000007801 UP000007819 UP000030742 UP000008711 UP000000803 UP000019118 UP000092553 UP000002282 UP000009192 UP000183832 UP000092443 UP000000673 UP000092445 UP000092461 UP000069272 UP000015103 UP000009046 UP000001070 UP000030765 UP000075884
UP000053105 UP000053825 UP000242457 UP000076502 UP000069940 UP000249989 UP000008820 UP000078542 UP000235965 UP000008237 UP000078541 UP000279307 UP000078492 UP000053097 UP000075809 UP000007755 UP000078540 UP000005205 UP000007266 UP000000311 UP000027135 UP000002358 UP000095301 UP000095300 UP000268350 UP000192223 UP000008792 UP000215335 UP000037069 UP000007801 UP000007819 UP000030742 UP000008711 UP000000803 UP000019118 UP000092553 UP000002282 UP000009192 UP000183832 UP000092443 UP000000673 UP000092445 UP000092461 UP000069272 UP000015103 UP000009046 UP000001070 UP000030765 UP000075884
PRIDE
Interpro
IPR000595
cNMP-bd_dom
+ More
IPR014710 RmlC-like_jellyroll
IPR000644 CBS_dom
IPR018490 cNMP-bd-like
IPR002550 CNNM
IPR001680 WD40_repeat
IPR014906 PRP4-like
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR036285 PRP4-like_sf
IPR020472 G-protein_beta_WD-40_rep
IPR014710 RmlC-like_jellyroll
IPR000644 CBS_dom
IPR018490 cNMP-bd-like
IPR002550 CNNM
IPR001680 WD40_repeat
IPR014906 PRP4-like
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR036285 PRP4-like_sf
IPR020472 G-protein_beta_WD-40_rep
Gene 3D
CDD
ProteinModelPortal
A0A2A4K7S3
A0A194QA38
A0A212F849
A0A3S2TFD0
H9JJ62
A0A2A4JS31
+ More
A0A2H1VKG4 A0A088AEY8 A0A0N0BJY2 A0A0L7QKM1 A0A2A3EE16 A0A310S7G1 A0A154PFC6 A0A1L8DDT5 A0A182GUV4 Q16HD8 A0A195C5G0 A0A2J7RPU8 A0A023EVD1 E2BD06 A0A195EXQ7 A0A3L8E0X0 E9J5L7 A0A151J4V4 A0A026WNK7 A0A151WQ63 F4WTD0 A0A195BB62 A0A158NVU0 A0A1Q3G0I8 D6W9C1 A0A1Q3G0K9 E2AJ07 A0A067QMK4 K7J1W6 A0A1I8NKL3 A0A1I8MXE5 A0A1I8PUF5 A0A1I8PUG6 T1PG40 A0A1B6H3Z8 A0A1B6G206 A0A1Y1JZ23 A0A3B0JTY0 A0A1W4XF17 A0A1B6FH96 A0A1B6F7C0 A0A0Q9WAQ1 A0A232FCI9 A0A2H8TPI9 A0A0L0BQR4 A0A0P8XKN0 W8B844 A0A2S2NWZ3 A0A1Y1JR54 J9JL81 A0A1Y1JU74 U4U987 A0A0Q5WLR7 A0A0B7P9G0 N6TAW9 A0A0M4EQT3 A0A034W374 A0A0K8W7G5 A0A0R1ED38 A0A0R1EAX5 A0A0R1E7R6 A0A0Q9X6C8 A0A1J1IK57 A0A1A9XGV8 A0A0A1WZ12 A0A1B6DP17 A0A2M4BBT6 A0A2M4BBM2 W5JKZ4 A0A2M3Z3Q6 A0A2M4BC05 A0A2M3ZGL8 A0A1B0A504 A0A2M4ABE8 A0A1B0GLB6 A0A2M4AB51 A0A2M4ABR6 A0A1B6DR42 A0A182FRV8 T1I5R7 A0A1B6KU35 A0A2M3Z561 E0VYD8 A0A2R7X6X1 B4JAK1 A0A146KNK0 A0A084WSG8 A0A0A9WHS3 A0A146LPK8 A0A182NFP3
A0A2H1VKG4 A0A088AEY8 A0A0N0BJY2 A0A0L7QKM1 A0A2A3EE16 A0A310S7G1 A0A154PFC6 A0A1L8DDT5 A0A182GUV4 Q16HD8 A0A195C5G0 A0A2J7RPU8 A0A023EVD1 E2BD06 A0A195EXQ7 A0A3L8E0X0 E9J5L7 A0A151J4V4 A0A026WNK7 A0A151WQ63 F4WTD0 A0A195BB62 A0A158NVU0 A0A1Q3G0I8 D6W9C1 A0A1Q3G0K9 E2AJ07 A0A067QMK4 K7J1W6 A0A1I8NKL3 A0A1I8MXE5 A0A1I8PUF5 A0A1I8PUG6 T1PG40 A0A1B6H3Z8 A0A1B6G206 A0A1Y1JZ23 A0A3B0JTY0 A0A1W4XF17 A0A1B6FH96 A0A1B6F7C0 A0A0Q9WAQ1 A0A232FCI9 A0A2H8TPI9 A0A0L0BQR4 A0A0P8XKN0 W8B844 A0A2S2NWZ3 A0A1Y1JR54 J9JL81 A0A1Y1JU74 U4U987 A0A0Q5WLR7 A0A0B7P9G0 N6TAW9 A0A0M4EQT3 A0A034W374 A0A0K8W7G5 A0A0R1ED38 A0A0R1EAX5 A0A0R1E7R6 A0A0Q9X6C8 A0A1J1IK57 A0A1A9XGV8 A0A0A1WZ12 A0A1B6DP17 A0A2M4BBT6 A0A2M4BBM2 W5JKZ4 A0A2M3Z3Q6 A0A2M4BC05 A0A2M3ZGL8 A0A1B0A504 A0A2M4ABE8 A0A1B0GLB6 A0A2M4AB51 A0A2M4ABR6 A0A1B6DR42 A0A182FRV8 T1I5R7 A0A1B6KU35 A0A2M3Z561 E0VYD8 A0A2R7X6X1 B4JAK1 A0A146KNK0 A0A084WSG8 A0A0A9WHS3 A0A146LPK8 A0A182NFP3
PDB
4IY0
E-value=5.27717e-55,
Score=546
Ontologies
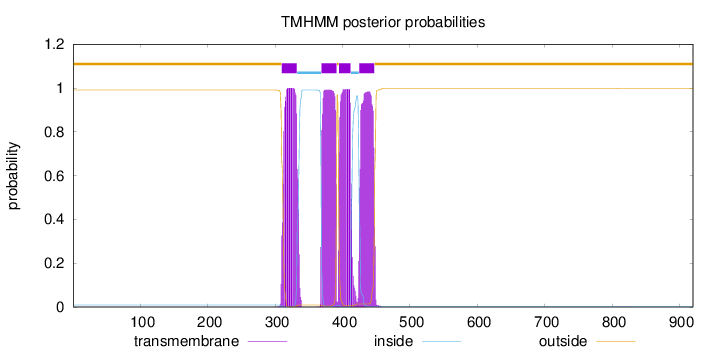
Topology
Length:
920
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
86.4901500000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00899
outside
1 - 309
TMhelix
310 - 332
inside
333 - 368
TMhelix
369 - 391
outside
392 - 394
TMhelix
395 - 412
inside
413 - 424
TMhelix
425 - 447
outside
448 - 920
Population Genetic Test Statistics
Pi
258.061895
Theta
188.580252
Tajima's D
1.373313
CLR
0.247002
CSRT
0.75921203939803
Interpretation
Uncertain
