Gene
KWMTBOMO14608 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008779
Annotation
PREDICTED:_putative_U5_small_nuclear_ribonucleoprotein_200_kDa_helicase_[Papilio_xuthus]
Full name
Putative U5 small nuclear ribonucleoprotein 200 kDa helicase
Alternative Name
Protein lethal (3) 72Ab
Location in the cell
Cytoplasmic Reliability : 1.08 Mitochondrial Reliability : 1.157
Sequence
CDS
ATGTCCGGGAGCTACTCGCTTGTGACTCATCTTAAACGCATCTCGTGCCCCCAGGTCATAAAGAAACTCGAGAAGAAGAACTTCCCGTGGGAGAAGCTGTACGAGCTGGGCCCCAACGAGATCGGCGAGCTGGTCCGCGCTCCGAAGCTGGGCAAGATGCTGCACAAGTACGTGCACCAGTTCCCGAAGCTGGAGCTCGCGACGCACATCCAGCCCATCACCAGGTCGACGCTCAGGGTCGAGCTCACCATCACGCCGGACTTCCATTGGGACGAGAAAATCCACGGACAGTCGGAGGCGTTCTGGATCCTGGTGGAGGACGTGGACTCTGAGGTGATCCTCCACCACGAGTATTTCCTGCTCAAGCAGAAGTACTGCAAGGACGAGCACCAGGTGAAGCTGTTCGTGCCGGTCTTCGAGCCCCTGCCTCCTCAGTACTTCCTGAGGGTCGTGTCCGATAGATGGATAGCAGCGGAGACGCAGCTTCCGGTGTCGTTCCGGCACCTCATCCTGCCCGAGAAGAACTTCCCGCCGACCGAGCTGCTGGACCTGCAGCCGCTGCCCATCTCGGCACTCCGGAATGCCAAGTACGAGGAGCTGTACAACGACACCTTCCCGCAGTTCAACCCGGTGCAGACGCAGGTGTTCAACGCAGTGTACAACTCTGACGACAACGTGTTCGTGGGTGCGCCGTCGGGCTCGGGCAAGTCGCTGGTTGCGGAGCTGGCGCTGCTGCGGATGCTGTCCCGGGACGCGGGCGGGCGCGCCGTGTACCTGCTGCCCAAGGACGCGCTGGCGGACATCGTGTTCGCGCACTGCTACCACAAGTTCGGGACCAAGTTCAATCTTAAGGTGGTCCAGCTGACCGGCGAGACTGCTACGGACCACAAGCTGCTCAACAAAGGCCAGATCATCATAACGACGGCCGATAAGTGGGACATACTCTCCCGGAGATGGAAAGTACGCAAGAGCGTCCAGAGCGTGTCGCTGCTGATCGCGGACGCGCTGCAGCTGCTGGGCGGCGAGGAGGGGCCGGTCGTGGAGGTGGTCTGCTCTAGGATGAGGTACATCGCCTCTCAGACAGGGAAGCCGATCCGTATAGTGGCCCTGTCGCTACCGCTGGCCGACGCCCGCGACGTTGCCCAGTGGCTCGGCTGCAACGCGAATGCCACCTTCAATTTCCACCCTAGCGTCAGATCCCTCCCACTAGAGTTGCACATCCAGGGCTTCAACATATCGCACGCGCCGACCCGGCTGACGGCCATGACGAAGCCCATCTACAACGCGATCCGGCGGCACGCGGGCACCCGCCCCTGCGCAGTGTTCGTGAGCTCGCGCCGCGCCGCCCGCCTGCTCGCCGCCGACCTGCTGGCGCTGGCGGCCGCGCGCGCAGCCCCCGCCGCCTTCCTGCACGCCTCGCGCGACCTGCTCGCGCCCTTCCTCGACGCCGTGCAGGACAAGATGCTGAAGGAGACGCTGTCCGCGGGCGTGGCGTACCTGCACGCGGGCGTGTCCGCCGGCGACCGGCGCGTGGTGCAGCAGCTGGTAGAGTGCGGCGCCGCGCAGCTGTGCGTGGTCGCCGCCGAGCTGGCCTGGAGCTTCGCCGCGCACGTGCACACCGTCATCGTGGCCGACACGTGCACGTACAACGGCAAGCTGCACTCGTACGAGCAGTACCCGATAACGACGGTGCTGCAGATGATCGGGCGCGCGTGCCGCCCGCTGGAGGACGACCACTCGGTGGCGGTGCTCATGTGCGCCGCGCACCACAAGACCTTCTTCACCAAGTTGCTCAACGACTGTCTGCCGCTGGAGAGTCACTTGGACCACAGACTCCACGATCACATGAACGCCGAGATAGTGACGAAGACCATAGAGAACAAGCAAGACGCCGTCGACTACTTGACCTGGACATTCCTGTACCGGCGGCTCACGCAGAACCCCAACTACTACAACTTACAGGGGGTCACGCACAGGCACCTGTCGGACCACCTCTCCGAGCTGGTGGAGACTACGCTGACGGATCTTGAGCAGTCCAAGTGCATCGCGATAGAGGATGAAATGGATGTCCAGCCACTGAACTTGGGGATGATAGCCTCATACTACTATATCAACTATACGACCATAGAACTGTTTAGTTTGTCTCTGACGAGCAAAACGAAGATCCGCGGCCTGCTCGAGATAATATCGTCGGCGGCCGAGTACTCGGACCTCTGCATCCGACACCGGGAGGAGAACATCATCAAATCTCTCGCCTCCAAGGTCCCGCACAAGCCGACGGCCCCGTCTGGCGGCGCCGTGCGGCACAACGACCCGCACGTGAAGGCGCACGTGCTGCTCCAGGCGCACCTCTCCAGGATGCAACTACCCGCCGAGCTGCAGGCCGACACCGCGCTCGTGCTCACCAAGGCGCTTCGGCTGATCCAGGCGTGCGTGGACGTGGTGTCCAGCAGCGGGTGGCTGGCGCCCGCCGTGGCCGCCATGGAGCTAGCCCAGATGGTGACGCAGGCCATGTGGGCCAAGGACTCGTACCTAAGGCAGCTGCCGCACTTCACGCCCGAGCTGGTGCAACGCTGCGCTGACAAGGGCGTGGAGACCGTGTTCGACGTGATGGAGCTGGAGGACACCGCCAGAGCCAAGCTGCTGCAGCTGTCTCCCGCCGAGATGGCAGACGTGGCGCGCTTCTGTAACCGGTACCCCAACGTGGAGCTCAACTATGAGGTGAAGAACGAGCGCCACCTGCGCGCCGGGAAGCCGATCGTGGTGGAGGTTTCGCTGGAGAGGGAGGACGACGTCCTCGGCCCGGTCATAGCGCCCTTCTTCCCCCAGAAACGCGAGGAGGGCTGGTGGGTCGTGATCGGAGACCCCAAGAGCAACACGCTGCTCTCCATCAAGCGGGTGTCGCTGGCCAGGAGCGCCAAGGTCCGGCTGGACTTCGTGTGTGGCGGCGCGGGGCGGCACACGTACACCCTGTACTTCATGTCCGACGCGTACCTAGGTGCCGACCAGGAGTACAAGTTCAACGTTGAGGTGGCAGACGCGGCGTCCGACTCCAGCGAGGACTCGGACCAGTACGGCTGA
Protein
MSGSYSLVTHLKRISCPQVIKKLEKKNFPWEKLYELGPNEIGELVRAPKLGKMLHKYVHQFPKLELATHIQPITRSTLRVELTITPDFHWDEKIHGQSEAFWILVEDVDSEVILHHEYFLLKQKYCKDEHQVKLFVPVFEPLPPQYFLRVVSDRWIAAETQLPVSFRHLILPEKNFPPTELLDLQPLPISALRNAKYEELYNDTFPQFNPVQTQVFNAVYNSDDNVFVGAPSGSGKSLVAELALLRMLSRDAGGRAVYLLPKDALADIVFAHCYHKFGTKFNLKVVQLTGETATDHKLLNKGQIIITTADKWDILSRRWKVRKSVQSVSLLIADALQLLGGEEGPVVEVVCSRMRYIASQTGKPIRIVALSLPLADARDVAQWLGCNANATFNFHPSVRSLPLELHIQGFNISHAPTRLTAMTKPIYNAIRRHAGTRPCAVFVSSRRAARLLAADLLALAAARAAPAAFLHASRDLLAPFLDAVQDKMLKETLSAGVAYLHAGVSAGDRRVVQQLVECGAAQLCVVAAELAWSFAAHVHTVIVADTCTYNGKLHSYEQYPITTVLQMIGRACRPLEDDHSVAVLMCAAHHKTFFTKLLNDCLPLESHLDHRLHDHMNAEIVTKTIENKQDAVDYLTWTFLYRRLTQNPNYYNLQGVTHRHLSDHLSELVETTLTDLEQSKCIAIEDEMDVQPLNLGMIASYYYINYTTIELFSLSLTSKTKIRGLLEIISSAAEYSDLCIRHREENIIKSLASKVPHKPTAPSGGAVRHNDPHVKAHVLLQAHLSRMQLPAELQADTALVLTKALRLIQACVDVVSSSGWLAPAVAAMELAQMVTQAMWAKDSYLRQLPHFTPELVQRCADKGVETVFDVMELEDTARAKLLQLSPAEMADVARFCNRYPNVELNYEVKNERHLRAGKPIVVEVSLEREDDVLGPVIAPFFPQKREEGWWVVIGDPKSNTLLSIKRVSLARSAKVRLDFVCGGAGRHTYTLYFMSDAYLGADQEYKFNVEVADAASDSSEDSDQYG
Summary
Description
Putative RNA helicase involved in the second step of RNA splicing.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the helicase family. SKI2 subfamily.
Keywords
ATP-binding
Complete proteome
Helicase
Hydrolase
mRNA processing
mRNA splicing
Nucleotide-binding
Nucleus
Reference proteome
Repeat
Spliceosome
Feature
chain Putative U5 small nuclear ribonucleoprotein 200 kDa helicase
Uniprot
A0A1W4WLT7
A0A139WNM0
A0A1I8MTD7
A0A1I8Q351
A0A1Y1MYR1
B0X1D8
+ More
T1PLJ4 A0A0A1WFR4 A0A0L0CI23 W8BT97 A0A182R4Y3 A0A0K8UJE3 A0A1Q3F5S4 A0A182VWG1 A0A182NQD8 A0A2J7RCH2 A0A182QXG8 A0A182YAH9 A0A182PGX5 A0A182MTI2 A0A084W518 A0A1S4FSY9 Q16QS5 A0A067RJY2 A0A182UQQ8 A0A182IVX8 A0A2J7RCH6 A0A336MEZ9 A0A182TM35 A0A182WS83 A0A182JU03 Q7PXQ1 A0A1Y9G9K8 A0A1B0FDY4 A0A182HL33 W5JH80 A0A1A9UPZ3 A0A1A9ZDE9 A0A182T6K2 A0A2M4A6R2 N6TX23 A0A1A9XFP2 A0A2P8YP35 A0A182H4Y4 B4KYI7 A0A1B0B7S0 A0A1A9WKU3 B4LDY9 A0A0M4EMC3 A0A2A3EKY9 A0A1B0D9F3 A0A1W4VP22 B3M9U8 B4J3P7 Q9VUV9 A0A0J9RW02 E9IWL3 A0A1B6C919 B3NDP3 F4WS28 E2B7U8 B4HIL1 A0A158NJE4 A0A195CCL5 B4PD16 B4N718 Q2M0I9 A0A026W729 A0A0J7KTK4 E2A6Q4 A0A3B0JVC3 A0A2J7RCH7 J9K3Y5 A0A087TVN5 A0A0L7QNR8 A0A0C9R8Q1 A0A0T6B4B5 U4U3Y2 A0A1S3IYW6 A0A023GME8 A0A1J1J3B5 A0A232EQ00 K7JA29 A0A131YJT3 L7M5R0 A0A1B0CJP7 A0A2R5LJE3 A0A224Z6E7 A0A1Z5KU77 A0A1E1X934 A0A226MD86 A0A3B3T1P8 A0A3B3T081 R0LG86 H2RSA0 A0A293MXH2 A0A3B3SZX8 A0A3B3T1E4 A0A1W5ALR0 A0A3Q3LQG7
T1PLJ4 A0A0A1WFR4 A0A0L0CI23 W8BT97 A0A182R4Y3 A0A0K8UJE3 A0A1Q3F5S4 A0A182VWG1 A0A182NQD8 A0A2J7RCH2 A0A182QXG8 A0A182YAH9 A0A182PGX5 A0A182MTI2 A0A084W518 A0A1S4FSY9 Q16QS5 A0A067RJY2 A0A182UQQ8 A0A182IVX8 A0A2J7RCH6 A0A336MEZ9 A0A182TM35 A0A182WS83 A0A182JU03 Q7PXQ1 A0A1Y9G9K8 A0A1B0FDY4 A0A182HL33 W5JH80 A0A1A9UPZ3 A0A1A9ZDE9 A0A182T6K2 A0A2M4A6R2 N6TX23 A0A1A9XFP2 A0A2P8YP35 A0A182H4Y4 B4KYI7 A0A1B0B7S0 A0A1A9WKU3 B4LDY9 A0A0M4EMC3 A0A2A3EKY9 A0A1B0D9F3 A0A1W4VP22 B3M9U8 B4J3P7 Q9VUV9 A0A0J9RW02 E9IWL3 A0A1B6C919 B3NDP3 F4WS28 E2B7U8 B4HIL1 A0A158NJE4 A0A195CCL5 B4PD16 B4N718 Q2M0I9 A0A026W729 A0A0J7KTK4 E2A6Q4 A0A3B0JVC3 A0A2J7RCH7 J9K3Y5 A0A087TVN5 A0A0L7QNR8 A0A0C9R8Q1 A0A0T6B4B5 U4U3Y2 A0A1S3IYW6 A0A023GME8 A0A1J1J3B5 A0A232EQ00 K7JA29 A0A131YJT3 L7M5R0 A0A1B0CJP7 A0A2R5LJE3 A0A224Z6E7 A0A1Z5KU77 A0A1E1X934 A0A226MD86 A0A3B3T1P8 A0A3B3T081 R0LG86 H2RSA0 A0A293MXH2 A0A3B3SZX8 A0A3B3T1E4 A0A1W5ALR0 A0A3Q3LQG7
EC Number
3.6.4.13
Pubmed
18362917
19820115
25315136
28004739
25830018
26108605
+ More
24495485 25244985 24438588 17510324 24845553 12364791 14747013 17210077 20920257 23761445 23537049 29403074 26483478 17994087 10731132 12537572 12537569 22936249 21282665 21719571 20798317 21347285 17550304 15632085 24508170 30249741 28648823 20075255 26830274 25576852 28797301 28528879 28503490 29240929 21551351
24495485 25244985 24438588 17510324 24845553 12364791 14747013 17210077 20920257 23761445 23537049 29403074 26483478 17994087 10731132 12537572 12537569 22936249 21282665 21719571 20798317 21347285 17550304 15632085 24508170 30249741 28648823 20075255 26830274 25576852 28797301 28528879 28503490 29240929 21551351
EMBL
KQ971308
KYB29542.1
GEZM01017312
JAV90779.1
DS232258
EDS38587.1
+ More
KA648950 AFP63579.1 GBXI01016997 JAC97294.1 JRES01000371 KNC31881.1 GAMC01014126 JAB92429.1 GDHF01025525 JAI26789.1 GFDL01012138 JAV22907.1 NEVH01005885 PNF38528.1 AXCN02001526 AXCM01000983 ATLV01020452 KE525302 KFB45312.1 CH477735 EAT36755.1 KK852636 KDR19736.1 PNF38530.1 UFQT01000821 SSX27373.1 AAAB01008987 EAA00850.3 CCAG010021136 APCN01000879 ADMH02001504 ETN62280.1 GGFK01003111 MBW36432.1 APGK01048231 APGK01048232 APGK01048233 APGK01048234 APGK01048235 APGK01048236 APGK01048237 APGK01048238 APGK01048239 KB741102 ENN73865.1 PYGN01000456 PSN46005.1 JXUM01110490 JXUM01110491 JXUM01110492 JXUM01110493 KQ565417 KXJ71002.1 CH933809 EDW18798.1 JXJN01009710 CH940647 EDW70032.1 CP012525 ALC44797.1 KZ288227 PBC31829.1 AJVK01028188 AJVK01028189 CH902618 EDV40139.1 CH916366 EDV97278.1 AE014296 BT016121 AY069270 CM002912 KMY99883.1 GL766582 EFZ15034.1 GEDC01027568 JAS09730.1 CH954178 EDV52176.1 GL888295 EGI63007.1 GL446209 EFN88241.1 CH480815 EDW41641.1 ADTU01017656 ADTU01017657 ADTU01017658 ADTU01017659 ADTU01017660 ADTU01017661 KQ977935 KYM98547.1 CM000159 EDW94948.2 CH964168 EDW80157.2 CH379069 EAL30943.2 KK107364 QOIP01000007 EZA51912.1 RLU20421.1 LBMM01003333 KMQ93648.1 GL437183 EFN70894.1 OUUW01000009 SPP85013.1 PNF38531.1 ABLF02033400 KK116951 KFM69174.1 KQ414845 KOC60268.1 GBYB01012739 JAG82506.1 LJIG01015965 KRT82021.1 KB631615 ERL84685.1 GBBM01000407 JAC35011.1 CVRI01000064 CRL05385.1 NNAY01002835 OXU20469.1 AAZX01012231 GEDV01009742 JAP78815.1 GACK01005418 JAA59616.1 AJWK01014826 AJWK01014827 GGLE01005488 MBY09614.1 GFPF01010756 MAA21902.1 GFJQ02008323 JAV98646.1 GFAC01003440 JAT95748.1 MCFN01001355 OXB53211.1 KB743182 EOB00570.1 GFWV01020794 MAA45522.1
KA648950 AFP63579.1 GBXI01016997 JAC97294.1 JRES01000371 KNC31881.1 GAMC01014126 JAB92429.1 GDHF01025525 JAI26789.1 GFDL01012138 JAV22907.1 NEVH01005885 PNF38528.1 AXCN02001526 AXCM01000983 ATLV01020452 KE525302 KFB45312.1 CH477735 EAT36755.1 KK852636 KDR19736.1 PNF38530.1 UFQT01000821 SSX27373.1 AAAB01008987 EAA00850.3 CCAG010021136 APCN01000879 ADMH02001504 ETN62280.1 GGFK01003111 MBW36432.1 APGK01048231 APGK01048232 APGK01048233 APGK01048234 APGK01048235 APGK01048236 APGK01048237 APGK01048238 APGK01048239 KB741102 ENN73865.1 PYGN01000456 PSN46005.1 JXUM01110490 JXUM01110491 JXUM01110492 JXUM01110493 KQ565417 KXJ71002.1 CH933809 EDW18798.1 JXJN01009710 CH940647 EDW70032.1 CP012525 ALC44797.1 KZ288227 PBC31829.1 AJVK01028188 AJVK01028189 CH902618 EDV40139.1 CH916366 EDV97278.1 AE014296 BT016121 AY069270 CM002912 KMY99883.1 GL766582 EFZ15034.1 GEDC01027568 JAS09730.1 CH954178 EDV52176.1 GL888295 EGI63007.1 GL446209 EFN88241.1 CH480815 EDW41641.1 ADTU01017656 ADTU01017657 ADTU01017658 ADTU01017659 ADTU01017660 ADTU01017661 KQ977935 KYM98547.1 CM000159 EDW94948.2 CH964168 EDW80157.2 CH379069 EAL30943.2 KK107364 QOIP01000007 EZA51912.1 RLU20421.1 LBMM01003333 KMQ93648.1 GL437183 EFN70894.1 OUUW01000009 SPP85013.1 PNF38531.1 ABLF02033400 KK116951 KFM69174.1 KQ414845 KOC60268.1 GBYB01012739 JAG82506.1 LJIG01015965 KRT82021.1 KB631615 ERL84685.1 GBBM01000407 JAC35011.1 CVRI01000064 CRL05385.1 NNAY01002835 OXU20469.1 AAZX01012231 GEDV01009742 JAP78815.1 GACK01005418 JAA59616.1 AJWK01014826 AJWK01014827 GGLE01005488 MBY09614.1 GFPF01010756 MAA21902.1 GFJQ02008323 JAV98646.1 GFAC01003440 JAT95748.1 MCFN01001355 OXB53211.1 KB743182 EOB00570.1 GFWV01020794 MAA45522.1
Proteomes
UP000192223
UP000007266
UP000095301
UP000095300
UP000002320
UP000037069
+ More
UP000075900 UP000075920 UP000075884 UP000235965 UP000075886 UP000076408 UP000075885 UP000075883 UP000030765 UP000008820 UP000027135 UP000075903 UP000075880 UP000075902 UP000076407 UP000075881 UP000007062 UP000069272 UP000092444 UP000075840 UP000000673 UP000078200 UP000092445 UP000075901 UP000019118 UP000092443 UP000245037 UP000069940 UP000249989 UP000009192 UP000092460 UP000091820 UP000008792 UP000092553 UP000242457 UP000092462 UP000192221 UP000007801 UP000001070 UP000000803 UP000008711 UP000007755 UP000008237 UP000001292 UP000005205 UP000078542 UP000002282 UP000007798 UP000001819 UP000053097 UP000279307 UP000036403 UP000000311 UP000268350 UP000007819 UP000054359 UP000053825 UP000030742 UP000085678 UP000183832 UP000215335 UP000002358 UP000092461 UP000198323 UP000261540 UP000005226 UP000192224 UP000261640
UP000075900 UP000075920 UP000075884 UP000235965 UP000075886 UP000076408 UP000075885 UP000075883 UP000030765 UP000008820 UP000027135 UP000075903 UP000075880 UP000075902 UP000076407 UP000075881 UP000007062 UP000069272 UP000092444 UP000075840 UP000000673 UP000078200 UP000092445 UP000075901 UP000019118 UP000092443 UP000245037 UP000069940 UP000249989 UP000009192 UP000092460 UP000091820 UP000008792 UP000092553 UP000242457 UP000092462 UP000192221 UP000007801 UP000001070 UP000000803 UP000008711 UP000007755 UP000008237 UP000001292 UP000005205 UP000078542 UP000002282 UP000007798 UP000001819 UP000053097 UP000279307 UP000036403 UP000000311 UP000268350 UP000007819 UP000054359 UP000053825 UP000030742 UP000085678 UP000183832 UP000215335 UP000002358 UP000092461 UP000198323 UP000261540 UP000005226 UP000192224 UP000261640
Interpro
IPR011545
DEAD/DEAH_box_helicase_dom
+ More
IPR035892 C2_domain_sf
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR036390 WH_DNA-bd_sf
IPR041094 Brr2_helicase_PWI
IPR003593 AAA+_ATPase
IPR027417 P-loop_NTPase
IPR014756 Ig_E-set
IPR004179 Sec63-dom
IPR003604 Matrin/U1-like-C_Znf_C2H2
IPR032259 HIBYL-CoA-H
IPR029045 ClpP/crotonase-like_dom_sf
IPR035892 C2_domain_sf
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR036390 WH_DNA-bd_sf
IPR041094 Brr2_helicase_PWI
IPR003593 AAA+_ATPase
IPR027417 P-loop_NTPase
IPR014756 Ig_E-set
IPR004179 Sec63-dom
IPR003604 Matrin/U1-like-C_Znf_C2H2
IPR032259 HIBYL-CoA-H
IPR029045 ClpP/crotonase-like_dom_sf
Gene 3D
CDD
ProteinModelPortal
A0A1W4WLT7
A0A139WNM0
A0A1I8MTD7
A0A1I8Q351
A0A1Y1MYR1
B0X1D8
+ More
T1PLJ4 A0A0A1WFR4 A0A0L0CI23 W8BT97 A0A182R4Y3 A0A0K8UJE3 A0A1Q3F5S4 A0A182VWG1 A0A182NQD8 A0A2J7RCH2 A0A182QXG8 A0A182YAH9 A0A182PGX5 A0A182MTI2 A0A084W518 A0A1S4FSY9 Q16QS5 A0A067RJY2 A0A182UQQ8 A0A182IVX8 A0A2J7RCH6 A0A336MEZ9 A0A182TM35 A0A182WS83 A0A182JU03 Q7PXQ1 A0A1Y9G9K8 A0A1B0FDY4 A0A182HL33 W5JH80 A0A1A9UPZ3 A0A1A9ZDE9 A0A182T6K2 A0A2M4A6R2 N6TX23 A0A1A9XFP2 A0A2P8YP35 A0A182H4Y4 B4KYI7 A0A1B0B7S0 A0A1A9WKU3 B4LDY9 A0A0M4EMC3 A0A2A3EKY9 A0A1B0D9F3 A0A1W4VP22 B3M9U8 B4J3P7 Q9VUV9 A0A0J9RW02 E9IWL3 A0A1B6C919 B3NDP3 F4WS28 E2B7U8 B4HIL1 A0A158NJE4 A0A195CCL5 B4PD16 B4N718 Q2M0I9 A0A026W729 A0A0J7KTK4 E2A6Q4 A0A3B0JVC3 A0A2J7RCH7 J9K3Y5 A0A087TVN5 A0A0L7QNR8 A0A0C9R8Q1 A0A0T6B4B5 U4U3Y2 A0A1S3IYW6 A0A023GME8 A0A1J1J3B5 A0A232EQ00 K7JA29 A0A131YJT3 L7M5R0 A0A1B0CJP7 A0A2R5LJE3 A0A224Z6E7 A0A1Z5KU77 A0A1E1X934 A0A226MD86 A0A3B3T1P8 A0A3B3T081 R0LG86 H2RSA0 A0A293MXH2 A0A3B3SZX8 A0A3B3T1E4 A0A1W5ALR0 A0A3Q3LQG7
T1PLJ4 A0A0A1WFR4 A0A0L0CI23 W8BT97 A0A182R4Y3 A0A0K8UJE3 A0A1Q3F5S4 A0A182VWG1 A0A182NQD8 A0A2J7RCH2 A0A182QXG8 A0A182YAH9 A0A182PGX5 A0A182MTI2 A0A084W518 A0A1S4FSY9 Q16QS5 A0A067RJY2 A0A182UQQ8 A0A182IVX8 A0A2J7RCH6 A0A336MEZ9 A0A182TM35 A0A182WS83 A0A182JU03 Q7PXQ1 A0A1Y9G9K8 A0A1B0FDY4 A0A182HL33 W5JH80 A0A1A9UPZ3 A0A1A9ZDE9 A0A182T6K2 A0A2M4A6R2 N6TX23 A0A1A9XFP2 A0A2P8YP35 A0A182H4Y4 B4KYI7 A0A1B0B7S0 A0A1A9WKU3 B4LDY9 A0A0M4EMC3 A0A2A3EKY9 A0A1B0D9F3 A0A1W4VP22 B3M9U8 B4J3P7 Q9VUV9 A0A0J9RW02 E9IWL3 A0A1B6C919 B3NDP3 F4WS28 E2B7U8 B4HIL1 A0A158NJE4 A0A195CCL5 B4PD16 B4N718 Q2M0I9 A0A026W729 A0A0J7KTK4 E2A6Q4 A0A3B0JVC3 A0A2J7RCH7 J9K3Y5 A0A087TVN5 A0A0L7QNR8 A0A0C9R8Q1 A0A0T6B4B5 U4U3Y2 A0A1S3IYW6 A0A023GME8 A0A1J1J3B5 A0A232EQ00 K7JA29 A0A131YJT3 L7M5R0 A0A1B0CJP7 A0A2R5LJE3 A0A224Z6E7 A0A1Z5KU77 A0A1E1X934 A0A226MD86 A0A3B3T1P8 A0A3B3T081 R0LG86 H2RSA0 A0A293MXH2 A0A3B3SZX8 A0A3B3T1E4 A0A1W5ALR0 A0A3Q3LQG7
PDB
6QX9
E-value=0,
Score=3602
Ontologies
GO
GO:0003676
GO:0004386
GO:0005524
GO:0046540
GO:0004004
GO:0000388
GO:0071013
GO:0071014
GO:0000398
GO:0005682
GO:0008026
GO:0005681
GO:0005634
GO:0008270
GO:0003860
GO:0016021
GO:0000381
GO:0030532
GO:0007283
GO:0007419
GO:0000278
GO:0071011
GO:0008380
GO:0003724
GO:0042802
GO:0005654
GO:0005515
GO:0003677
GO:0006101
GO:0003824
GO:0008152
GO:0016491
GO:0015930
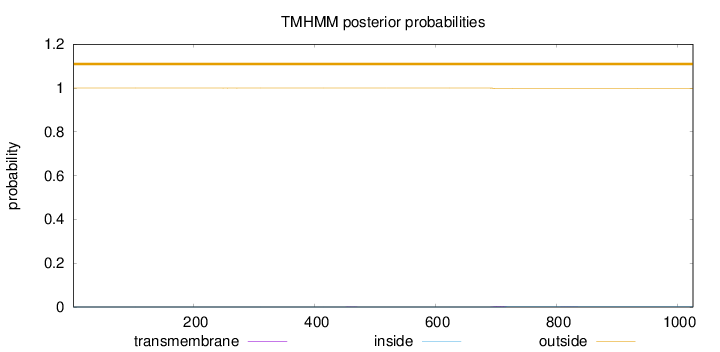
Topology
Subcellular location
Nucleus
Length:
1026
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0573300000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00017
outside
1 - 1026
Population Genetic Test Statistics
Pi
255.519491
Theta
175.522474
Tajima's D
1.838676
CLR
0.212611
CSRT
0.849357532123394
Interpretation
Uncertain
