Gene
KWMTBOMO14571
Pre Gene Modal
BGIBMGA008737
Annotation
PREDICTED:_protein_TEX261_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.926
Sequence
CDS
ATGATATTCGAGGAGATCCCCCTCCAGATGAACATCCTAGGTCTGATACAGCAGCTTCTGCACCTGCTTCTGCTGCGGGACTTCCCCGCGGTCCGGGTGACCAGCTTCAGTTTCTTGACTGCACTGATGACGCTCGCGGTTCATCATTATCTGGCCTTCAAGTACTTCGGAGCGGTGTACTACGCTTTTTCGGAGGTGCTGGCCTACTTCACGCTCTGTCTATGGGTGGTTCCCTTCGGTCTTTTCGTGTCCCTCTCAGCAAACGACTACGTGCTACCAACGACAGAAGAGAAGCACCCGCTGTTGGGTGACAGTAACGTTCTGACAGATTATCTGTCGCGTAAATCGAAAAAATACAGTCTACTCTCGTTCTTCAGTTACGCCAAGGAGTCCATTCTACCTCACCGGAACAAGAAAGCGTTCTGA
Protein
MIFEEIPLQMNILGLIQQLLHLLLLRDFPAVRVTSFSFLTALMTLAVHHYLAFKYFGAVYYAFSEVLAYFTLCLWVVPFGLFVSLSANDYVLPTTEEKHPLLGDSNVLTDYLSRKSKKYSLLSFFSYAKESILPHRNKKAF
Summary
Uniprot
A0A2A4K7C5
A0A2H1VRI9
A0A0N1PHJ2
A0A212ESP7
A0A0N1ID45
A0A2H1VCL5
+ More
A0A3S2LRN8 A0A2J7Q085 A0A2P8XDU0 A0A1B6DTA1 A0A1B6EF10 A0A067QVS3 A0A182YPF7 A0A1B6GUG7 A0A1B6GR63 A0A1B6I163 A0A1B6KPM9 U5EY91 T1JH68 A0A182LWU6 A0A182R8Q2 A0A2M4AM10 A0A2M3ZAG8 A0A182WF57 W5JSZ1 A0A2M4AM35 A0A1Q3EUB5 A0A2M4CQT8 A0A2M4CQC4 T1D4X6 A0A1L8DJW5 A0A2P6KIN2 A0A182JWF3 A0A182P9C8 A0A2R5LD45 A0A2L2XZF6 A0A293MY37 Q7Q331 A0A182XCA3 A0A182LPI0 A0A182TFL5 A0A182HNK4 A0A182VF72 Q171I0 A0A023ELB2 Q16PH9 B7PM75 A0A0K8RQL4 A0A182MXW4 A0A087UTR6 A0A182QFG0 A0A023FMS3 A0A084WBE6 A0A0C9RXW1 A0A023FFI5 G3MML7 L7M525 A0A023GMM6 A0A131YT13 A0A023FX73 A0A1I8N764 A0A0A1WMR9 A0A226E2R3 A0A1B6CZ52 B4G9Y9 A0A1J1HXR0 B4MNH8 A0A0L8FNZ7 C1BN86 A0A0U2MAG9 Q292A6 A0A2C9LU98 A0A3B0J2Y7 C1BR71 A0A0K8VVY2 A0A139WAM0 A0A1I8Q5A4 A0A3S1BTF9 A0A1S3KEY5 A0A1W4WWD4 B4LPM3 A0A1W4VR87 T1KNE6 V4AD90 D3PJS4 R4G8M3 K1PGP2 A0A1A9ZZB0 A0A1B0FDI2 A0A1A9V3B7 A0A0L0BXH3 A0A1D2NLQ5 A0A1V9Y3I5 B3S905 A0A0T6AZH1 A0A2R7X144
A0A3S2LRN8 A0A2J7Q085 A0A2P8XDU0 A0A1B6DTA1 A0A1B6EF10 A0A067QVS3 A0A182YPF7 A0A1B6GUG7 A0A1B6GR63 A0A1B6I163 A0A1B6KPM9 U5EY91 T1JH68 A0A182LWU6 A0A182R8Q2 A0A2M4AM10 A0A2M3ZAG8 A0A182WF57 W5JSZ1 A0A2M4AM35 A0A1Q3EUB5 A0A2M4CQT8 A0A2M4CQC4 T1D4X6 A0A1L8DJW5 A0A2P6KIN2 A0A182JWF3 A0A182P9C8 A0A2R5LD45 A0A2L2XZF6 A0A293MY37 Q7Q331 A0A182XCA3 A0A182LPI0 A0A182TFL5 A0A182HNK4 A0A182VF72 Q171I0 A0A023ELB2 Q16PH9 B7PM75 A0A0K8RQL4 A0A182MXW4 A0A087UTR6 A0A182QFG0 A0A023FMS3 A0A084WBE6 A0A0C9RXW1 A0A023FFI5 G3MML7 L7M525 A0A023GMM6 A0A131YT13 A0A023FX73 A0A1I8N764 A0A0A1WMR9 A0A226E2R3 A0A1B6CZ52 B4G9Y9 A0A1J1HXR0 B4MNH8 A0A0L8FNZ7 C1BN86 A0A0U2MAG9 Q292A6 A0A2C9LU98 A0A3B0J2Y7 C1BR71 A0A0K8VVY2 A0A139WAM0 A0A1I8Q5A4 A0A3S1BTF9 A0A1S3KEY5 A0A1W4WWD4 B4LPM3 A0A1W4VR87 T1KNE6 V4AD90 D3PJS4 R4G8M3 K1PGP2 A0A1A9ZZB0 A0A1B0FDI2 A0A1A9V3B7 A0A0L0BXH3 A0A1D2NLQ5 A0A1V9Y3I5 B3S905 A0A0T6AZH1 A0A2R7X144
Pubmed
26354079
22118469
29403074
24845553
25244985
20920257
+ More
23761445 24330624 26561354 12364791 14747013 17210077 20966253 17510324 24945155 26483478 24438588 26131772 22216098 25576852 26830274 25315136 25830018 17994087 26621068 15632085 15562597 18362917 19820115 23254933 22992520 26108605 27289101 28327890 18719581
23761445 24330624 26561354 12364791 14747013 17210077 20966253 17510324 24945155 26483478 24438588 26131772 22216098 25576852 26830274 25315136 25830018 17994087 26621068 15632085 15562597 18362917 19820115 23254933 22992520 26108605 27289101 28327890 18719581
EMBL
NWSH01000101
PCG79552.1
ODYU01003993
SOQ43417.1
KQ459805
KPJ19895.1
+ More
AGBW02012775 OWR44474.1 KQ459348 KPJ01313.1 ODYU01001824 SOQ38555.1 RSAL01000335 RVE42462.1 NEVH01019998 PNF21995.1 PYGN01002626 PSN30170.1 GEDC01008400 JAS28898.1 GEDC01000775 JAS36523.1 KK853313 KDR08618.1 GECZ01003703 JAS66066.1 GECZ01004861 JAS64908.1 GECU01027038 JAS80668.1 GEBQ01026569 JAT13408.1 GANO01000512 JAB59359.1 JH432222 AXCM01000254 GGFK01008498 MBW41819.1 GGFM01004697 MBW25448.1 ADMH02000540 ETN66050.1 GGFK01008481 MBW41802.1 GFDL01016136 JAV18909.1 GGFL01003363 MBW67541.1 GGFL01003364 MBW67542.1 GALA01000793 JAA94059.1 GFDF01007326 JAV06758.1 MWRG01010989 PRD26196.1 GGLE01003324 MBY07450.1 IAAA01008523 LAA00225.1 GFWV01020120 MAA44848.1 AAAB01008966 EAA13015.1 APCN01001939 CH477454 EAT40652.1 JXUM01147162 GAPW01004564 KQ570310 JAC09034.1 KXJ68391.1 CH477781 EAT36269.1 ABJB010396851 ABJB010408345 DS746366 EEC07697.1 GADI01000457 JAA73351.1 KK121574 KFM80755.1 AXCN02002114 GBBK01001928 JAC22554.1 ATLV01022347 KE525331 KFB47540.1 GBZX01000259 JAG92481.1 GBBK01004622 JAC19860.1 JO843118 AEO34735.1 GACK01006067 JAA58967.1 GBBM01000324 JAC35094.1 GEDV01006819 JAP81738.1 GBBL01001972 JAC25348.1 GBXI01014569 JAC99722.1 LNIX01000007 OXA51719.1 GEDC01018653 JAS18645.1 CH479181 EDW31741.1 CVRI01000024 CRK92124.1 CH963848 EDW73667.1 KQ428101 KOF66441.1 BT076065 ACO10489.1 KT755309 ALS05143.1 CM000071 EAL24956.1 OUUW01000001 SPP73562.1 BT077100 ACO11524.1 GDHF01009609 JAI42705.1 KQ971382 KYB24941.1 RQTK01000022 RUS90914.1 CH940648 EDW60261.1 CAEY01000276 KB201977 ESO93075.1 BT121880 ADD38810.1 ACPB03020017 GAHY01000722 JAA76788.1 JH818308 EKC18039.1 CCAG010008713 JRES01001190 KNC24691.1 LJIJ01000009 ODN06218.1 MNPL01000194 OQR80243.1 DS985257 EDV20862.1 LJIG01022458 KRT80474.1 KK856330 PTY25518.1
AGBW02012775 OWR44474.1 KQ459348 KPJ01313.1 ODYU01001824 SOQ38555.1 RSAL01000335 RVE42462.1 NEVH01019998 PNF21995.1 PYGN01002626 PSN30170.1 GEDC01008400 JAS28898.1 GEDC01000775 JAS36523.1 KK853313 KDR08618.1 GECZ01003703 JAS66066.1 GECZ01004861 JAS64908.1 GECU01027038 JAS80668.1 GEBQ01026569 JAT13408.1 GANO01000512 JAB59359.1 JH432222 AXCM01000254 GGFK01008498 MBW41819.1 GGFM01004697 MBW25448.1 ADMH02000540 ETN66050.1 GGFK01008481 MBW41802.1 GFDL01016136 JAV18909.1 GGFL01003363 MBW67541.1 GGFL01003364 MBW67542.1 GALA01000793 JAA94059.1 GFDF01007326 JAV06758.1 MWRG01010989 PRD26196.1 GGLE01003324 MBY07450.1 IAAA01008523 LAA00225.1 GFWV01020120 MAA44848.1 AAAB01008966 EAA13015.1 APCN01001939 CH477454 EAT40652.1 JXUM01147162 GAPW01004564 KQ570310 JAC09034.1 KXJ68391.1 CH477781 EAT36269.1 ABJB010396851 ABJB010408345 DS746366 EEC07697.1 GADI01000457 JAA73351.1 KK121574 KFM80755.1 AXCN02002114 GBBK01001928 JAC22554.1 ATLV01022347 KE525331 KFB47540.1 GBZX01000259 JAG92481.1 GBBK01004622 JAC19860.1 JO843118 AEO34735.1 GACK01006067 JAA58967.1 GBBM01000324 JAC35094.1 GEDV01006819 JAP81738.1 GBBL01001972 JAC25348.1 GBXI01014569 JAC99722.1 LNIX01000007 OXA51719.1 GEDC01018653 JAS18645.1 CH479181 EDW31741.1 CVRI01000024 CRK92124.1 CH963848 EDW73667.1 KQ428101 KOF66441.1 BT076065 ACO10489.1 KT755309 ALS05143.1 CM000071 EAL24956.1 OUUW01000001 SPP73562.1 BT077100 ACO11524.1 GDHF01009609 JAI42705.1 KQ971382 KYB24941.1 RQTK01000022 RUS90914.1 CH940648 EDW60261.1 CAEY01000276 KB201977 ESO93075.1 BT121880 ADD38810.1 ACPB03020017 GAHY01000722 JAA76788.1 JH818308 EKC18039.1 CCAG010008713 JRES01001190 KNC24691.1 LJIJ01000009 ODN06218.1 MNPL01000194 OQR80243.1 DS985257 EDV20862.1 LJIG01022458 KRT80474.1 KK856330 PTY25518.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000283053
UP000235965
+ More
UP000245037 UP000027135 UP000076408 UP000075883 UP000075900 UP000075920 UP000000673 UP000075881 UP000075885 UP000007062 UP000076407 UP000075882 UP000075902 UP000075840 UP000075903 UP000008820 UP000069940 UP000249989 UP000001555 UP000075884 UP000054359 UP000075886 UP000030765 UP000095301 UP000198287 UP000008744 UP000183832 UP000007798 UP000053454 UP000001819 UP000076420 UP000268350 UP000007266 UP000095300 UP000271974 UP000085678 UP000192223 UP000008792 UP000192221 UP000015104 UP000030746 UP000015103 UP000005408 UP000092445 UP000092444 UP000078200 UP000037069 UP000094527 UP000192247 UP000009022
UP000245037 UP000027135 UP000076408 UP000075883 UP000075900 UP000075920 UP000000673 UP000075881 UP000075885 UP000007062 UP000076407 UP000075882 UP000075902 UP000075840 UP000075903 UP000008820 UP000069940 UP000249989 UP000001555 UP000075884 UP000054359 UP000075886 UP000030765 UP000095301 UP000198287 UP000008744 UP000183832 UP000007798 UP000053454 UP000001819 UP000076420 UP000268350 UP000007266 UP000095300 UP000271974 UP000085678 UP000192223 UP000008792 UP000192221 UP000015104 UP000030746 UP000015103 UP000005408 UP000092445 UP000092444 UP000078200 UP000037069 UP000094527 UP000192247 UP000009022
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A2A4K7C5
A0A2H1VRI9
A0A0N1PHJ2
A0A212ESP7
A0A0N1ID45
A0A2H1VCL5
+ More
A0A3S2LRN8 A0A2J7Q085 A0A2P8XDU0 A0A1B6DTA1 A0A1B6EF10 A0A067QVS3 A0A182YPF7 A0A1B6GUG7 A0A1B6GR63 A0A1B6I163 A0A1B6KPM9 U5EY91 T1JH68 A0A182LWU6 A0A182R8Q2 A0A2M4AM10 A0A2M3ZAG8 A0A182WF57 W5JSZ1 A0A2M4AM35 A0A1Q3EUB5 A0A2M4CQT8 A0A2M4CQC4 T1D4X6 A0A1L8DJW5 A0A2P6KIN2 A0A182JWF3 A0A182P9C8 A0A2R5LD45 A0A2L2XZF6 A0A293MY37 Q7Q331 A0A182XCA3 A0A182LPI0 A0A182TFL5 A0A182HNK4 A0A182VF72 Q171I0 A0A023ELB2 Q16PH9 B7PM75 A0A0K8RQL4 A0A182MXW4 A0A087UTR6 A0A182QFG0 A0A023FMS3 A0A084WBE6 A0A0C9RXW1 A0A023FFI5 G3MML7 L7M525 A0A023GMM6 A0A131YT13 A0A023FX73 A0A1I8N764 A0A0A1WMR9 A0A226E2R3 A0A1B6CZ52 B4G9Y9 A0A1J1HXR0 B4MNH8 A0A0L8FNZ7 C1BN86 A0A0U2MAG9 Q292A6 A0A2C9LU98 A0A3B0J2Y7 C1BR71 A0A0K8VVY2 A0A139WAM0 A0A1I8Q5A4 A0A3S1BTF9 A0A1S3KEY5 A0A1W4WWD4 B4LPM3 A0A1W4VR87 T1KNE6 V4AD90 D3PJS4 R4G8M3 K1PGP2 A0A1A9ZZB0 A0A1B0FDI2 A0A1A9V3B7 A0A0L0BXH3 A0A1D2NLQ5 A0A1V9Y3I5 B3S905 A0A0T6AZH1 A0A2R7X144
A0A3S2LRN8 A0A2J7Q085 A0A2P8XDU0 A0A1B6DTA1 A0A1B6EF10 A0A067QVS3 A0A182YPF7 A0A1B6GUG7 A0A1B6GR63 A0A1B6I163 A0A1B6KPM9 U5EY91 T1JH68 A0A182LWU6 A0A182R8Q2 A0A2M4AM10 A0A2M3ZAG8 A0A182WF57 W5JSZ1 A0A2M4AM35 A0A1Q3EUB5 A0A2M4CQT8 A0A2M4CQC4 T1D4X6 A0A1L8DJW5 A0A2P6KIN2 A0A182JWF3 A0A182P9C8 A0A2R5LD45 A0A2L2XZF6 A0A293MY37 Q7Q331 A0A182XCA3 A0A182LPI0 A0A182TFL5 A0A182HNK4 A0A182VF72 Q171I0 A0A023ELB2 Q16PH9 B7PM75 A0A0K8RQL4 A0A182MXW4 A0A087UTR6 A0A182QFG0 A0A023FMS3 A0A084WBE6 A0A0C9RXW1 A0A023FFI5 G3MML7 L7M525 A0A023GMM6 A0A131YT13 A0A023FX73 A0A1I8N764 A0A0A1WMR9 A0A226E2R3 A0A1B6CZ52 B4G9Y9 A0A1J1HXR0 B4MNH8 A0A0L8FNZ7 C1BN86 A0A0U2MAG9 Q292A6 A0A2C9LU98 A0A3B0J2Y7 C1BR71 A0A0K8VVY2 A0A139WAM0 A0A1I8Q5A4 A0A3S1BTF9 A0A1S3KEY5 A0A1W4WWD4 B4LPM3 A0A1W4VR87 T1KNE6 V4AD90 D3PJS4 R4G8M3 K1PGP2 A0A1A9ZZB0 A0A1B0FDI2 A0A1A9V3B7 A0A0L0BXH3 A0A1D2NLQ5 A0A1V9Y3I5 B3S905 A0A0T6AZH1 A0A2R7X144
Ontologies
PANTHER
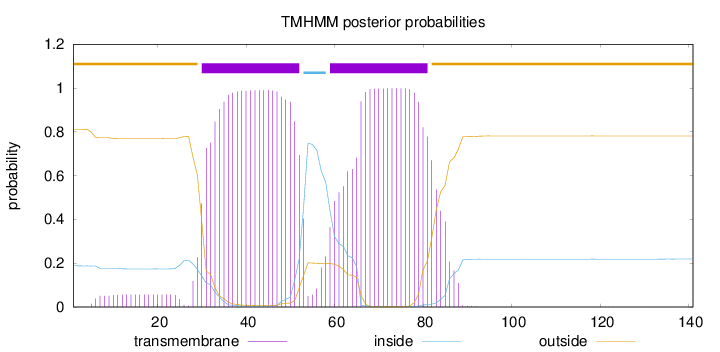
Topology
Length:
141
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.11401
Exp number, first 60 AAs:
24.15608
Total prob of N-in:
0.18866
POSSIBLE N-term signal
sequence
outside
1 - 29
TMhelix
30 - 52
inside
53 - 58
TMhelix
59 - 81
outside
82 - 141
Population Genetic Test Statistics
Pi
256.048453
Theta
169.129223
Tajima's D
1.973161
CLR
0.21491
CSRT
0.88075596220189
Interpretation
Uncertain
