Gene
KWMTBOMO14562
Pre Gene Modal
BGIBMGA012878
Annotation
PREDICTED:_uncharacterized_protein_LOC106710540_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.295
Sequence
CDS
ATGGAGCGGCCCGGAACTAGCGGGGCCAGCACTCCGGTCATAGAGCGGCCCGGAACTAGCGGGGCCAGCACCCTGGTCATGGAGCGGCCTGGCACTAGCGAGGCCAGCACCTCGGCCACCGAGCGGCCTGTATCCTCGAGGCCATCTTATGGCGATGACGCTGTAAGTTATGTGCAGCTGAAACGCGAAGGAAGTCTGTGCACCGTCAAAGCAAAACTTTGCCCTGAGCATAAGGTCCACGCAAAATTGTACTTAGTAACACTTATAGTCGACGAAGAGGATGAAGTAGTGAAATCTGTGCAATGCCATGGCTGTGTTGCTTCGCAAGGAGGCTGCAAACATGCTGTGGCTTTTCTCATGTGGGTCCATCGCCGAAGTGAGGAGCCTTCCTGTACATCTGTGGAGTGCTACTGGAGAAAATCAAAGCTATCAGGTGTTGGTACCACAATAAAATATATAACTTCTAAACAATTATCTAATGGAAAACCAAGCTTACCATCAAATAATGGTGTGTTTACAAAATTTATAGAAGAAGGGAAGAAACGTAAGCTTAGTGACTGTGAGGTACTTAAGTATCAAGCAGACTATTTTCCTGATACATTAGAAAGATTATCAATGCATAAACTGGTACAGAAATGCAAAGAAAAATCCTGTGACACCTTTTTTGAAAAAGTTGAGTTAACTAATGAAGATATTATAAAAGTTGAGGAAGCAACTAGAGAACAACATAAAAGCTCGCTTTGGTATGAATTGAGGTATAGCAGAGTGACAGCTTCACGAGCTTTTGAATTCAGTAGGTGCAAAACCACTGATAGCACCTTGATTTCGTTGATAATGGGAGGGAAAATTCCAGATACATGGGCCATGAAGCACGGTAGAATGTTGGAGAATGAAGTGAGGCAAACAGTAAGCAGCATACTGAGAAAAAAATAA
Protein
MERPGTSGASTPVIERPGTSGASTLVMERPGTSEASTSATERPVSSRPSYGDDAVSYVQLKREGSLCTVKAKLCPEHKVHAKLYLVTLIVDEEDEVVKSVQCHGCVASQGGCKHAVAFLMWVHRRSEEPSCTSVECYWRKSKLSGVGTTIKYITSKQLSNGKPSLPSNNGVFTKFIEEGKKRKLSDCEVLKYQADYFPDTLERLSMHKLVQKCKEKSCDTFFEKVELTNEDIIKVEEATREQHKSSLWYELRYSRVTASRAFEFSRCKTTDSTLISLIMGGKIPDTWAMKHGRMLENEVRQTVSSILRKK
Summary
Uniprot
A0A0L7LJE3
A0A0L7KYC4
A0A2A4K405
A0A3S2LJ79
A0A2A4J2K2
A0A2W1BN93
+ More
A0A2H1VDK3 A0A3S2NZG9 A0A0L7L459 A0A1W4WCR0 J9KSL0 A0A3S2M7J6 K7JM84 A0A1S3CX42 A0A1S3D3I6 K7JUJ0 A0A3Q0IM63 A0A164KWR3 A0A1S3D9C1 A0A164TFV3 A0A195EE43 J9JYB7 A0A164NZ12 A0A151IFX9 E9HHA3 A0A151I6S0 A0A1S4EQI7 A0A151J732 A0A151JMW2 E9HXR3 A0A151IWY7 A0A146MFR2 A0A151J6P5 A0A151JQL6 B0XI73 B0XEB5 A0A182UE76 A0A182VAA1 A0A182HWP6 A0A182PHQ3 A0A182LIZ6 A0A182XMU0 Q7Q6T3 Q17PM9 A0A182UJA7 A0A182GDQ2 A0A182WIF3 A0A1Q3F618 A0A1Q3F586 A0A182HDS0 A0A182K9V7 A0A182MSJ5 A0A151IJL4 A0A151I6J3 A0A182RL91 A0A182S7C4 A0A182QJ89 A0A182Y1C8 A0A182NGZ2 A0A2M4BMV0 A0A2M4BR26 A0A2M3ZBL9 W5J6F7
A0A2H1VDK3 A0A3S2NZG9 A0A0L7L459 A0A1W4WCR0 J9KSL0 A0A3S2M7J6 K7JM84 A0A1S3CX42 A0A1S3D3I6 K7JUJ0 A0A3Q0IM63 A0A164KWR3 A0A1S3D9C1 A0A164TFV3 A0A195EE43 J9JYB7 A0A164NZ12 A0A151IFX9 E9HHA3 A0A151I6S0 A0A1S4EQI7 A0A151J732 A0A151JMW2 E9HXR3 A0A151IWY7 A0A146MFR2 A0A151J6P5 A0A151JQL6 B0XI73 B0XEB5 A0A182UE76 A0A182VAA1 A0A182HWP6 A0A182PHQ3 A0A182LIZ6 A0A182XMU0 Q7Q6T3 Q17PM9 A0A182UJA7 A0A182GDQ2 A0A182WIF3 A0A1Q3F618 A0A1Q3F586 A0A182HDS0 A0A182K9V7 A0A182MSJ5 A0A151IJL4 A0A151I6J3 A0A182RL91 A0A182S7C4 A0A182QJ89 A0A182Y1C8 A0A182NGZ2 A0A2M4BMV0 A0A2M4BR26 A0A2M3ZBL9 W5J6F7
Pubmed
EMBL
JTDY01000906
KOB75489.1
JTDY01004524
KOB68049.1
NWSH01000174
PCG78756.1
+ More
RSAL01000095 RVE47830.1 NWSH01003931 PCG65670.1 KZ149984 PZC75741.1 ODYU01001989 SOQ38940.1 RSAL01000082 RVE48496.1 JTDY01003038 KOB70278.1 ABLF02004610 ABLF02004611 ABLF02004612 RSAL01000014 RVE53270.1 AAZX01003835 AAZX01004060 AAZX01005068 AAZX01005855 AAZX01006160 AAZX01003550 AAZX01011933 AAZX01014557 LRGB01003249 KZS03599.1 LRGB01001804 KZS10419.1 KQ979074 KYN23114.1 ABLF02015734 LRGB01002769 KZS06367.1 KQ977751 KYN00124.1 GL732647 EFX68881.1 KQ978464 KYM93794.1 KQ979748 KYN19357.1 KQ978888 KYN27669.1 GL733065 EFX63466.1 KQ980841 KYN12328.1 GDHC01000330 JAQ18299.1 KQ979820 KYN18945.1 KQ978690 KYN29277.1 DS233273 EDS29063.1 DS232815 EDS25889.1 APCN01001522 AAAB01008960 EAA11483.3 CH477190 EAT48722.1 EAT48723.1 JXUM01056320 KQ561905 KXJ77212.1 GFDL01012048 JAV22997.1 GFDL01012314 JAV22731.1 JXUM01130224 KQ567551 KXJ69379.1 AXCM01009087 KQ977397 KYN02978.1 KQ978476 KYM93678.1 AXCN02000350 GGFJ01005268 MBW54409.1 GGFJ01006292 MBW55433.1 GGFM01005205 MBW25956.1 ADMH02002072 ETN59586.1
RSAL01000095 RVE47830.1 NWSH01003931 PCG65670.1 KZ149984 PZC75741.1 ODYU01001989 SOQ38940.1 RSAL01000082 RVE48496.1 JTDY01003038 KOB70278.1 ABLF02004610 ABLF02004611 ABLF02004612 RSAL01000014 RVE53270.1 AAZX01003835 AAZX01004060 AAZX01005068 AAZX01005855 AAZX01006160 AAZX01003550 AAZX01011933 AAZX01014557 LRGB01003249 KZS03599.1 LRGB01001804 KZS10419.1 KQ979074 KYN23114.1 ABLF02015734 LRGB01002769 KZS06367.1 KQ977751 KYN00124.1 GL732647 EFX68881.1 KQ978464 KYM93794.1 KQ979748 KYN19357.1 KQ978888 KYN27669.1 GL733065 EFX63466.1 KQ980841 KYN12328.1 GDHC01000330 JAQ18299.1 KQ979820 KYN18945.1 KQ978690 KYN29277.1 DS233273 EDS29063.1 DS232815 EDS25889.1 APCN01001522 AAAB01008960 EAA11483.3 CH477190 EAT48722.1 EAT48723.1 JXUM01056320 KQ561905 KXJ77212.1 GFDL01012048 JAV22997.1 GFDL01012314 JAV22731.1 JXUM01130224 KQ567551 KXJ69379.1 AXCM01009087 KQ977397 KYN02978.1 KQ978476 KYM93678.1 AXCN02000350 GGFJ01005268 MBW54409.1 GGFJ01006292 MBW55433.1 GGFM01005205 MBW25956.1 ADMH02002072 ETN59586.1
Proteomes
UP000037510
UP000218220
UP000283053
UP000192223
UP000007819
UP000002358
+ More
UP000079169 UP000076858 UP000078492 UP000078542 UP000000305 UP000002320 UP000075902 UP000075903 UP000075840 UP000075885 UP000075882 UP000076407 UP000007062 UP000008820 UP000069940 UP000249989 UP000075920 UP000075881 UP000075883 UP000075900 UP000075901 UP000075886 UP000076408 UP000075884 UP000000673
UP000079169 UP000076858 UP000078492 UP000078542 UP000000305 UP000002320 UP000075902 UP000075903 UP000075840 UP000075885 UP000075882 UP000076407 UP000007062 UP000008820 UP000069940 UP000249989 UP000075920 UP000075881 UP000075883 UP000075900 UP000075901 UP000075886 UP000076408 UP000075884 UP000000673
Interpro
SUPFAM
SSF52980
SSF52980
Gene 3D
ProteinModelPortal
A0A0L7LJE3
A0A0L7KYC4
A0A2A4K405
A0A3S2LJ79
A0A2A4J2K2
A0A2W1BN93
+ More
A0A2H1VDK3 A0A3S2NZG9 A0A0L7L459 A0A1W4WCR0 J9KSL0 A0A3S2M7J6 K7JM84 A0A1S3CX42 A0A1S3D3I6 K7JUJ0 A0A3Q0IM63 A0A164KWR3 A0A1S3D9C1 A0A164TFV3 A0A195EE43 J9JYB7 A0A164NZ12 A0A151IFX9 E9HHA3 A0A151I6S0 A0A1S4EQI7 A0A151J732 A0A151JMW2 E9HXR3 A0A151IWY7 A0A146MFR2 A0A151J6P5 A0A151JQL6 B0XI73 B0XEB5 A0A182UE76 A0A182VAA1 A0A182HWP6 A0A182PHQ3 A0A182LIZ6 A0A182XMU0 Q7Q6T3 Q17PM9 A0A182UJA7 A0A182GDQ2 A0A182WIF3 A0A1Q3F618 A0A1Q3F586 A0A182HDS0 A0A182K9V7 A0A182MSJ5 A0A151IJL4 A0A151I6J3 A0A182RL91 A0A182S7C4 A0A182QJ89 A0A182Y1C8 A0A182NGZ2 A0A2M4BMV0 A0A2M4BR26 A0A2M3ZBL9 W5J6F7
A0A2H1VDK3 A0A3S2NZG9 A0A0L7L459 A0A1W4WCR0 J9KSL0 A0A3S2M7J6 K7JM84 A0A1S3CX42 A0A1S3D3I6 K7JUJ0 A0A3Q0IM63 A0A164KWR3 A0A1S3D9C1 A0A164TFV3 A0A195EE43 J9JYB7 A0A164NZ12 A0A151IFX9 E9HHA3 A0A151I6S0 A0A1S4EQI7 A0A151J732 A0A151JMW2 E9HXR3 A0A151IWY7 A0A146MFR2 A0A151J6P5 A0A151JQL6 B0XI73 B0XEB5 A0A182UE76 A0A182VAA1 A0A182HWP6 A0A182PHQ3 A0A182LIZ6 A0A182XMU0 Q7Q6T3 Q17PM9 A0A182UJA7 A0A182GDQ2 A0A182WIF3 A0A1Q3F618 A0A1Q3F586 A0A182HDS0 A0A182K9V7 A0A182MSJ5 A0A151IJL4 A0A151I6J3 A0A182RL91 A0A182S7C4 A0A182QJ89 A0A182Y1C8 A0A182NGZ2 A0A2M4BMV0 A0A2M4BR26 A0A2M3ZBL9 W5J6F7
Ontologies
GO
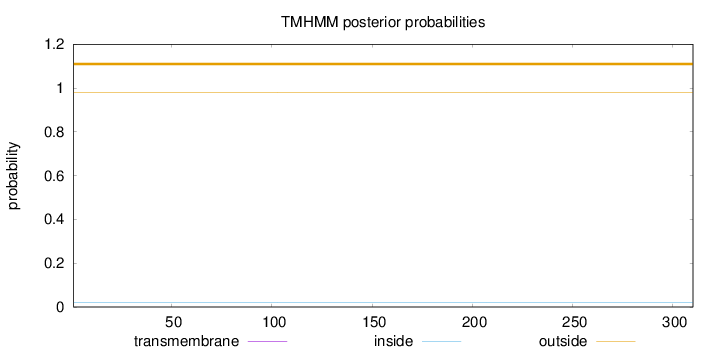
Topology
Length:
310
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00113
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02199
outside
1 - 310
Population Genetic Test Statistics
Pi
529.69512
Theta
238.00761
Tajima's D
3.835336
CLR
0.294267
CSRT
0.99830008499575
Interpretation
Uncertain
