Gene
KWMTBOMO14558
Annotation
retroelement_polyprotein_[Glyptapanteles_flavicoxis]
Location in the cell
Nuclear Reliability : 1.157 Peroxisomal Reliability : 1.094
Sequence
CDS
ATGTGTGTAGACTACCGAGAGTTAAATAGGAATACCGTCAGGGACCATTATGCCTTACCGATAATCTCAGACCAAATAGACCAACTGGCCGGGGGTATTTATTTCACCACAATTGACATGGCAGCTGGGTTTCATCAGATACCTATTTCGGAGGGATCAATTGAAAAGACTGCTTTTGTAACTCCCGATGGTTTATATGAATACCTAACTATGCCATTTGGACTTTGCAACGCGGTGTCAGTCTACCAGCGCTGCATTAATAGGGCTCTCGCTCACCTCACGAGCTCACCAGACCAAGTGAGCCAGGTCTATGTCGATGATGTGCTCTCAAAGTGTAAAGATTTTGATCAGGGCTTATACCATATTGAACGCATATTGATTGCTCTACAAGAAGCTGGCTTTTCTATTAACGTTGATAAAACCGCCTTCTTTAAACGCTCCATCGAGTACTTGGGCAATATAATTGCAGACGGGCAGGTCAGCCCTAGCCCAAGAAAGGTGGAGGCTTTAACGAAGGCACCAATCCCGAAAACTGTCAAACAAGTTAGACAATTCAATGGCCTGGCTGGATATTTCAGACGTTTCATTCCCAACTTCTCGCGAGTTATGGCTCCTTTATATGAACTCACTAAGAACGATACGACACGGGAATGGAACGAAAGGCATGACGAAGCTAGGGATTATATTGTCAAACATCTGTCTACTGCGCCTACCCTTACGCTATTCCAGGAAGACGCACCCATCGAACTCTACACGGACGCCAGCAGCATCGGCTACGGGGCAGTGTTAGTGCAGACATTCGCGGGTCGCCAACATCCAGTCGCGTACATGAGCCAGCGCACCACCGACGCCGAGAGCCGCTACCACTCTTACGAACTTGAAACGTTGGCGGTCGTAAGAGCAATTAAGCATTTTCGGCATTACCTCTACGGCCGAAAATTCAAGGTAATAACCGACTGTAATGCTCTCAAGGCCTCCAAAAACAAAAAGGACTTGCTTCCGCGAATCCATCGGTGGTGGGCGTTTTTGCAAAACTACGAATTCGAGGTGGAGTATAGGAAAGGTGAGCGATTACAGCACGCACTTTTTCAGTAG
Protein
MCVDYRELNRNTVRDHYALPIISDQIDQLAGGIYFTTIDMAAGFHQIPISEGSIEKTAFVTPDGLYEYLTMPFGLCNAVSVYQRCINRALAHLTSSPDQVSQVYVDDVLSKCKDFDQGLYHIERILIALQEAGFSINVDKTAFFKRSIEYLGNIIADGQVSPSPRKVEALTKAPIPKTVKQVRQFNGLAGYFRRFIPNFSRVMAPLYELTKNDTTREWNERHDEARDYIVKHLSTAPTLTLFQEDAPIELYTDASSIGYGAVLVQTFAGRQHPVAYMSQRTTDAESRYHSYELETLAVVRAIKHFRHYLYGRKFKVITDCNALKASKNKKDLLPRIHRWWAFLQNYEFEVEYRKGERLQHALFQ
Summary
Uniprot
A0A0A9WYE4
A0A146L2V0
A0A0A9W4E8
B7S8P8
A0A0A9WUU5
A0A0A9WTW7
+ More
A0A0A9Z954 A0A3S2L135 A0A146M2J2 A0A0A9X1Z8 A0A0A9WJJ8 V5GN60 A0A1Y1NG42 A0A1Y1NAV9 D2A5M1 A0A1E1VY25 Q24310 A0A0J7KEC9 A0A1Y1KFN9 A0A1Y1KFM9 A0A139WIB9 A0A0A9ZIH0 A0A0A9WB80 A0A0A9WGV0 A0A1E1W482 A0A0A9XL77 V5H1P2 A0A0A9YZ15 A0A0C9RXL6 A0A0A9ZDJ9 A0A139WJA1 A0A139WIQ0 A0A139WA12 A0A139WJA0 A0A139WIZ4 A0A2S2PFS2 A0A146KXF1 A0A1Y1NEJ5 A0A2S2R3T0 A0A2S2QVZ0 A0A2S2P032 A0A0J7KVJ0 A0A194R1L3 X1X5H2 A0A2W1BPU0 A0A146LDG4 X1XU22 A0A2L2Y402 A0A2L2Y1Z9 A0A2L2Y1I8 X1WPP0 A0A146KYH2 A0A2W1BA98 A0A1W7R6G2 A0A2S2PGQ9 A0A0A9YM49 J9JJ45 A0A147BLC5 J9JZ77 A0A164MP09 A0A1Y1LN89 A0A0A1XB16 X1XM84 A0A0A9YEP8 A0A023EY26 J9KKW9 J9JN68 A0A0J7K890 X1XD76 A0A2S2PMX3 A0A0J7NF11 A0A2S2NKJ2 J9K544 X1X261 A0A2H8TRV6 A0A0J7K9X8 A0A0J7KLK4 J9LBS9 A0A0A9XJ85 A0A1Z5L674 Q94885 X1XU14 A0A293LZX6 A0A023EZG2 A0A023EYH9 A0A023F013 A0A023EYF0 A0A1Y1MFA9 A0A0P6D622 A0A2S2NFQ4 A0A162R894 A0A164P0I2 A0A1Y1MFE0 A0A1Z5KVF1 A0A139W8S4 A0A162SB45 A0A1W7R6L8 A0A1W7R6G1 A0A0P5JUZ2
A0A0A9Z954 A0A3S2L135 A0A146M2J2 A0A0A9X1Z8 A0A0A9WJJ8 V5GN60 A0A1Y1NG42 A0A1Y1NAV9 D2A5M1 A0A1E1VY25 Q24310 A0A0J7KEC9 A0A1Y1KFN9 A0A1Y1KFM9 A0A139WIB9 A0A0A9ZIH0 A0A0A9WB80 A0A0A9WGV0 A0A1E1W482 A0A0A9XL77 V5H1P2 A0A0A9YZ15 A0A0C9RXL6 A0A0A9ZDJ9 A0A139WJA1 A0A139WIQ0 A0A139WA12 A0A139WJA0 A0A139WIZ4 A0A2S2PFS2 A0A146KXF1 A0A1Y1NEJ5 A0A2S2R3T0 A0A2S2QVZ0 A0A2S2P032 A0A0J7KVJ0 A0A194R1L3 X1X5H2 A0A2W1BPU0 A0A146LDG4 X1XU22 A0A2L2Y402 A0A2L2Y1Z9 A0A2L2Y1I8 X1WPP0 A0A146KYH2 A0A2W1BA98 A0A1W7R6G2 A0A2S2PGQ9 A0A0A9YM49 J9JJ45 A0A147BLC5 J9JZ77 A0A164MP09 A0A1Y1LN89 A0A0A1XB16 X1XM84 A0A0A9YEP8 A0A023EY26 J9KKW9 J9JN68 A0A0J7K890 X1XD76 A0A2S2PMX3 A0A0J7NF11 A0A2S2NKJ2 J9K544 X1X261 A0A2H8TRV6 A0A0J7K9X8 A0A0J7KLK4 J9LBS9 A0A0A9XJ85 A0A1Z5L674 Q94885 X1XU14 A0A293LZX6 A0A023EZG2 A0A023EYH9 A0A023F013 A0A023EYF0 A0A1Y1MFA9 A0A0P6D622 A0A2S2NFQ4 A0A162R894 A0A164P0I2 A0A1Y1MFE0 A0A1Z5KVF1 A0A139W8S4 A0A162SB45 A0A1W7R6L8 A0A1W7R6G1 A0A0P5JUZ2
Pubmed
EMBL
GBHO01040014
GBHO01031163
JAG03590.1
JAG12441.1
GDHC01016400
JAQ02229.1
+ More
GBHO01041313 JAG02291.1 EF710649 ACE75273.1 GBHO01032413 JAG11191.1 GBHO01032415 JAG11189.1 GBHO01040010 GBHO01001812 JAG03594.1 JAG41792.1 RSAL01000274 RVE43148.1 GDHC01004980 JAQ13649.1 GBHO01030786 JAG12818.1 GBHO01036023 GBHO01032007 JAG07581.1 JAG11597.1 GALX01002922 JAB65544.1 GEZM01007614 GEZM01007613 GEZM01007612 JAV95056.1 GEZM01007616 GEZM01007615 GEZM01007611 JAV95051.1 KQ971345 EFA05054.2 GDQN01011434 JAT79620.1 X14037 CAA32198.1 LBMM01008871 KMQ88569.1 GEZM01089268 GEZM01089261 JAV57577.1 GEZM01089269 GEZM01089265 JAV57567.1 KQ971339 KYB27733.1 GBHO01041187 GBHO01040948 GBHO01039116 GBHO01000859 JAG02417.1 JAG02656.1 JAG04488.1 JAG42745.1 GBHO01041099 GBHO01037897 JAG02505.1 JAG05707.1 GBHO01037851 JAG05753.1 GDQN01009269 JAT81785.1 GBHO01043544 GBHO01031946 GBHO01031931 GBHO01022910 GBHO01022909 GBHO01015498 GBHO01001135 GBHO01001106 JAG00060.1 JAG11658.1 JAG11673.1 JAG20694.1 JAG20695.1 JAG28106.1 JAG42469.1 JAG42498.1 GALX01001723 JAB66743.1 GBHO01022918 GBHO01006165 GBHO01006163 GBHO01001110 JAG20686.1 JAG37439.1 JAG37441.1 JAG42494.1 GBYB01013920 JAG83687.1 GBHO01041008 GBHO01006162 GBHO01001134 GBHO01001105 JAG02596.1 JAG37442.1 JAG42470.1 JAG42499.1 KQ971338 KYB27887.1 KYB27888.1 KQ971701 KYB24754.1 KYB27885.1 KYB27886.1 GGMR01015682 MBY28301.1 GDHC01018907 JAP99721.1 GEZM01005427 JAV96179.1 GGMS01015422 MBY84625.1 GGMS01012726 MBY81929.1 GGMR01009949 MBY22568.1 LBMM01002816 KMQ94326.1 KQ460883 KPJ11409.1 ABLF02004401 ABLF02013544 ABLF02013550 KZ149989 PZC75624.1 GDHC01012341 JAQ06288.1 ABLF02017401 IAAA01004057 LAA02040.1 IAAA01004056 LAA02037.1 IAAA01004058 LAA02042.1 ABLF02033943 ABLF02042737 GDHC01018562 JAQ00067.1 KZ150428 PZC70914.1 GEHC01000888 JAV46757.1 GGMR01016004 MBY28623.1 GBHO01011083 GBHO01009497 JAG32521.1 JAG34107.1 ABLF02008401 ABLF02008404 GEGO01004299 JAR91105.1 ABLF02014660 ABLF02042774 LRGB01002992 KZS05228.1 GEZM01051217 GEZM01051215 JAV75083.1 GBXI01005773 JAD08519.1 ABLF02004526 ABLF02054969 GBHO01015604 JAG28000.1 GBBI01004477 JAC14235.1 ABLF02013955 ABLF02009677 ABLF02021755 ABLF02021756 ABLF02021757 ABLF02021758 ABLF02021759 ABLF02021760 ABLF02022031 ABLF02041876 LBMM01011782 KMQ86633.1 ABLF02022032 GGMR01018192 MBY30811.1 LBMM01005919 KMQ91095.1 GGMR01005091 MBY17710.1 ABLF02023666 ABLF02005127 ABLF02005129 ABLF02007401 GFXV01005120 MBW16925.1 LBMM01010976 KMQ87102.1 KMQ91096.1 GBHO01026469 JAG17135.1 GFJQ02004121 JAW02849.1 X95908 CAA65152.1 ABLF02003120 ABLF02004015 ABLF02057179 ABLF02066208 GFWV01007660 MAA32390.1 GBBI01004551 JAC14161.1 GBBI01004550 JAC14162.1 GBBI01004613 JAC14099.1 GBBI01004549 JAC14163.1 GEZM01033083 JAV84424.1 GDIP01005668 JAM98047.1 GGMR01003388 MBY16007.1 LRGB01000231 KZS20305.1 LRGB01002748 KZS06406.1 GEZM01034557 JAV83728.1 GFJQ02008169 JAV98800.1 KQ973017 KXZ75691.1 LRGB01000072 KZS21146.1 GEHC01000930 JAV46715.1 GEHC01000891 JAV46754.1 GDIQ01198582 JAK53143.1
GBHO01041313 JAG02291.1 EF710649 ACE75273.1 GBHO01032413 JAG11191.1 GBHO01032415 JAG11189.1 GBHO01040010 GBHO01001812 JAG03594.1 JAG41792.1 RSAL01000274 RVE43148.1 GDHC01004980 JAQ13649.1 GBHO01030786 JAG12818.1 GBHO01036023 GBHO01032007 JAG07581.1 JAG11597.1 GALX01002922 JAB65544.1 GEZM01007614 GEZM01007613 GEZM01007612 JAV95056.1 GEZM01007616 GEZM01007615 GEZM01007611 JAV95051.1 KQ971345 EFA05054.2 GDQN01011434 JAT79620.1 X14037 CAA32198.1 LBMM01008871 KMQ88569.1 GEZM01089268 GEZM01089261 JAV57577.1 GEZM01089269 GEZM01089265 JAV57567.1 KQ971339 KYB27733.1 GBHO01041187 GBHO01040948 GBHO01039116 GBHO01000859 JAG02417.1 JAG02656.1 JAG04488.1 JAG42745.1 GBHO01041099 GBHO01037897 JAG02505.1 JAG05707.1 GBHO01037851 JAG05753.1 GDQN01009269 JAT81785.1 GBHO01043544 GBHO01031946 GBHO01031931 GBHO01022910 GBHO01022909 GBHO01015498 GBHO01001135 GBHO01001106 JAG00060.1 JAG11658.1 JAG11673.1 JAG20694.1 JAG20695.1 JAG28106.1 JAG42469.1 JAG42498.1 GALX01001723 JAB66743.1 GBHO01022918 GBHO01006165 GBHO01006163 GBHO01001110 JAG20686.1 JAG37439.1 JAG37441.1 JAG42494.1 GBYB01013920 JAG83687.1 GBHO01041008 GBHO01006162 GBHO01001134 GBHO01001105 JAG02596.1 JAG37442.1 JAG42470.1 JAG42499.1 KQ971338 KYB27887.1 KYB27888.1 KQ971701 KYB24754.1 KYB27885.1 KYB27886.1 GGMR01015682 MBY28301.1 GDHC01018907 JAP99721.1 GEZM01005427 JAV96179.1 GGMS01015422 MBY84625.1 GGMS01012726 MBY81929.1 GGMR01009949 MBY22568.1 LBMM01002816 KMQ94326.1 KQ460883 KPJ11409.1 ABLF02004401 ABLF02013544 ABLF02013550 KZ149989 PZC75624.1 GDHC01012341 JAQ06288.1 ABLF02017401 IAAA01004057 LAA02040.1 IAAA01004056 LAA02037.1 IAAA01004058 LAA02042.1 ABLF02033943 ABLF02042737 GDHC01018562 JAQ00067.1 KZ150428 PZC70914.1 GEHC01000888 JAV46757.1 GGMR01016004 MBY28623.1 GBHO01011083 GBHO01009497 JAG32521.1 JAG34107.1 ABLF02008401 ABLF02008404 GEGO01004299 JAR91105.1 ABLF02014660 ABLF02042774 LRGB01002992 KZS05228.1 GEZM01051217 GEZM01051215 JAV75083.1 GBXI01005773 JAD08519.1 ABLF02004526 ABLF02054969 GBHO01015604 JAG28000.1 GBBI01004477 JAC14235.1 ABLF02013955 ABLF02009677 ABLF02021755 ABLF02021756 ABLF02021757 ABLF02021758 ABLF02021759 ABLF02021760 ABLF02022031 ABLF02041876 LBMM01011782 KMQ86633.1 ABLF02022032 GGMR01018192 MBY30811.1 LBMM01005919 KMQ91095.1 GGMR01005091 MBY17710.1 ABLF02023666 ABLF02005127 ABLF02005129 ABLF02007401 GFXV01005120 MBW16925.1 LBMM01010976 KMQ87102.1 KMQ91096.1 GBHO01026469 JAG17135.1 GFJQ02004121 JAW02849.1 X95908 CAA65152.1 ABLF02003120 ABLF02004015 ABLF02057179 ABLF02066208 GFWV01007660 MAA32390.1 GBBI01004551 JAC14161.1 GBBI01004550 JAC14162.1 GBBI01004613 JAC14099.1 GBBI01004549 JAC14163.1 GEZM01033083 JAV84424.1 GDIP01005668 JAM98047.1 GGMR01003388 MBY16007.1 LRGB01000231 KZS20305.1 LRGB01002748 KZS06406.1 GEZM01034557 JAV83728.1 GFJQ02008169 JAV98800.1 KQ973017 KXZ75691.1 LRGB01000072 KZS21146.1 GEHC01000930 JAV46715.1 GEHC01000891 JAV46754.1 GDIQ01198582 JAK53143.1
Proteomes
Pfam
Interpro
IPR041373
RT_RNaseH
+ More
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR021109 Peptidase_aspartic_dom_sf
IPR036397 RNaseH_sf
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR018061 Retropepsins
IPR034122 Retropepsin-like_bacterial
IPR025836 Zn_knuckle_CX2CX4HX4C
IPR005162 Retrotrans_gag_dom
IPR028002 Myb_DNA-bind_5
IPR000331 Rap_GAP_dom
IPR035974 Rap/Ran-GAP_sf
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR032071 DUF4806
IPR019103 Peptidase_aspartic_DDI1-type
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR021109 Peptidase_aspartic_dom_sf
IPR036397 RNaseH_sf
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR018061 Retropepsins
IPR034122 Retropepsin-like_bacterial
IPR025836 Zn_knuckle_CX2CX4HX4C
IPR005162 Retrotrans_gag_dom
IPR028002 Myb_DNA-bind_5
IPR000331 Rap_GAP_dom
IPR035974 Rap/Ran-GAP_sf
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR032071 DUF4806
IPR019103 Peptidase_aspartic_DDI1-type
SUPFAM
ProteinModelPortal
A0A0A9WYE4
A0A146L2V0
A0A0A9W4E8
B7S8P8
A0A0A9WUU5
A0A0A9WTW7
+ More
A0A0A9Z954 A0A3S2L135 A0A146M2J2 A0A0A9X1Z8 A0A0A9WJJ8 V5GN60 A0A1Y1NG42 A0A1Y1NAV9 D2A5M1 A0A1E1VY25 Q24310 A0A0J7KEC9 A0A1Y1KFN9 A0A1Y1KFM9 A0A139WIB9 A0A0A9ZIH0 A0A0A9WB80 A0A0A9WGV0 A0A1E1W482 A0A0A9XL77 V5H1P2 A0A0A9YZ15 A0A0C9RXL6 A0A0A9ZDJ9 A0A139WJA1 A0A139WIQ0 A0A139WA12 A0A139WJA0 A0A139WIZ4 A0A2S2PFS2 A0A146KXF1 A0A1Y1NEJ5 A0A2S2R3T0 A0A2S2QVZ0 A0A2S2P032 A0A0J7KVJ0 A0A194R1L3 X1X5H2 A0A2W1BPU0 A0A146LDG4 X1XU22 A0A2L2Y402 A0A2L2Y1Z9 A0A2L2Y1I8 X1WPP0 A0A146KYH2 A0A2W1BA98 A0A1W7R6G2 A0A2S2PGQ9 A0A0A9YM49 J9JJ45 A0A147BLC5 J9JZ77 A0A164MP09 A0A1Y1LN89 A0A0A1XB16 X1XM84 A0A0A9YEP8 A0A023EY26 J9KKW9 J9JN68 A0A0J7K890 X1XD76 A0A2S2PMX3 A0A0J7NF11 A0A2S2NKJ2 J9K544 X1X261 A0A2H8TRV6 A0A0J7K9X8 A0A0J7KLK4 J9LBS9 A0A0A9XJ85 A0A1Z5L674 Q94885 X1XU14 A0A293LZX6 A0A023EZG2 A0A023EYH9 A0A023F013 A0A023EYF0 A0A1Y1MFA9 A0A0P6D622 A0A2S2NFQ4 A0A162R894 A0A164P0I2 A0A1Y1MFE0 A0A1Z5KVF1 A0A139W8S4 A0A162SB45 A0A1W7R6L8 A0A1W7R6G1 A0A0P5JUZ2
A0A0A9Z954 A0A3S2L135 A0A146M2J2 A0A0A9X1Z8 A0A0A9WJJ8 V5GN60 A0A1Y1NG42 A0A1Y1NAV9 D2A5M1 A0A1E1VY25 Q24310 A0A0J7KEC9 A0A1Y1KFN9 A0A1Y1KFM9 A0A139WIB9 A0A0A9ZIH0 A0A0A9WB80 A0A0A9WGV0 A0A1E1W482 A0A0A9XL77 V5H1P2 A0A0A9YZ15 A0A0C9RXL6 A0A0A9ZDJ9 A0A139WJA1 A0A139WIQ0 A0A139WA12 A0A139WJA0 A0A139WIZ4 A0A2S2PFS2 A0A146KXF1 A0A1Y1NEJ5 A0A2S2R3T0 A0A2S2QVZ0 A0A2S2P032 A0A0J7KVJ0 A0A194R1L3 X1X5H2 A0A2W1BPU0 A0A146LDG4 X1XU22 A0A2L2Y402 A0A2L2Y1Z9 A0A2L2Y1I8 X1WPP0 A0A146KYH2 A0A2W1BA98 A0A1W7R6G2 A0A2S2PGQ9 A0A0A9YM49 J9JJ45 A0A147BLC5 J9JZ77 A0A164MP09 A0A1Y1LN89 A0A0A1XB16 X1XM84 A0A0A9YEP8 A0A023EY26 J9KKW9 J9JN68 A0A0J7K890 X1XD76 A0A2S2PMX3 A0A0J7NF11 A0A2S2NKJ2 J9K544 X1X261 A0A2H8TRV6 A0A0J7K9X8 A0A0J7KLK4 J9LBS9 A0A0A9XJ85 A0A1Z5L674 Q94885 X1XU14 A0A293LZX6 A0A023EZG2 A0A023EYH9 A0A023F013 A0A023EYF0 A0A1Y1MFA9 A0A0P6D622 A0A2S2NFQ4 A0A162R894 A0A164P0I2 A0A1Y1MFE0 A0A1Z5KVF1 A0A139W8S4 A0A162SB45 A0A1W7R6L8 A0A1W7R6G1 A0A0P5JUZ2
PDB
4OL8
E-value=5.36618e-59,
Score=576
Ontologies
KEGG
GO
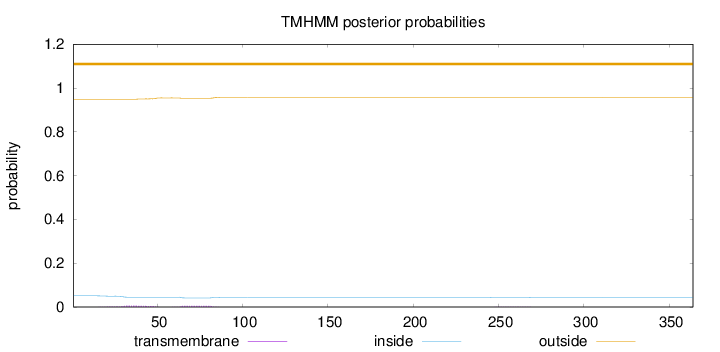
Topology
Length:
364
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23388
Exp number, first 60 AAs:
0.12524
Total prob of N-in:
0.05112
outside
1 - 364
Population Genetic Test Statistics
Pi
169.43453
Theta
74.850642
Tajima's D
2.584294
CLR
0.196249
CSRT
0.953452327383631
Interpretation
Uncertain
