Gene
KWMTBOMO14552
Pre Gene Modal
BGIBMGA013783
Annotation
AF461149_1_transposase_[Bombyx_mori]
Full name
Signal transducer and activator of transcription
Location in the cell
Mitochondrial Reliability : 2.127 Nuclear Reliability : 2.205
Sequence
CDS
ATGAAGACTAGTGTAGATGACAGGTCAAGAAGTGGTCGCCCTCGGTCTGTTAGGACTCCAGCAGTGATAAAAGCTGTGAAGGCGCGAATTCAAAGAAATCCCAAACGTAAGCAGAAATTGTTGGCCCTTCAGATGGGGTTAAGCAGAACCACGGTGAAAAGGGTGTTAAATGAAGACTTAGGGCTTCGGGCATATCGAAGAAAAACAGGACATCGTTTGAATGCTCGTCTAATGGACCTGAGACTGAAGAGATGCCGCGCTTTGTTGAAGCGGCACTGGGTATTCCAACAAGATTCGGCGCCAGCTCATAGAGCGAAGAGCACACAAGACTGGCTGGCGGCGCGTGAAATCGACTTCATCCGGCACGAAGACTGGCCCTCCTCCAGTCCAGATTTGAATCCGTTAGATTACAAGATATGGCAACACTTGGAGGAAAAGGCGTGCTCAAAGCCTCATCCCAATTTGGAGTCACTCAAGACATCCTTGATTAAGGCAGCCGCCGATATTGACATGGACCTCGTTCGTGCTGCGATAGACGACTGGCCGCGCAGATTGAAGGCCTGTATTCAAAATCACGGAGGTCATTTTGAATAA
Protein
MKTSVDDRSRSGRPRSVRTPAVIKAVKARIQRNPKRKQKLLALQMGLSRTTVKRVLNEDLGLRAYRRKTGHRLNARLMDLRLKRCRALLKRHWVFQQDSAPAHRAKSTQDWLAAREIDFIRHEDWPSSSPDLNPLDYKIWQHLEEKACSKPHPNLESLKTSLIKAAADIDMDLVRAAIDDWPRRLKACIQNHGGHFE
Summary
Similarity
Belongs to the transcription factor STAT family.
Belongs to the TGF-beta family.
Belongs to the TGF-beta family.
Uniprot
EMBL
Proteomes
Pfam
Interpro
IPR009057
Homeobox-like_sf
+ More
IPR012345 STAT_TF_DNA-bd_N
IPR008967 p53-like_TF_DNA-bd
IPR013799 STAT_TF_prot_interaction
IPR013800 STAT_TF_alpha
IPR013801 STAT_TF_DNA-bd
IPR000980 SH2
IPR015988 STAT_TF_coiled-coil
IPR001217 STAT
IPR036535 STAT_N_sf
IPR036860 SH2_dom_sf
IPR001839 TGF-b_C
IPR029034 Cystine-knot_cytokine
IPR017948 TGFb_CS
IPR015615 TGF-beta-rel
IPR032821 KAsynt_C_assoc
IPR014031 Ketoacyl_synth_C
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR016039 Thiolase-like
IPR014030 Ketoacyl_synth_N
IPR038717 Tc1-like_DDE_dom
IPR012345 STAT_TF_DNA-bd_N
IPR008967 p53-like_TF_DNA-bd
IPR013799 STAT_TF_prot_interaction
IPR013800 STAT_TF_alpha
IPR013801 STAT_TF_DNA-bd
IPR000980 SH2
IPR015988 STAT_TF_coiled-coil
IPR001217 STAT
IPR036535 STAT_N_sf
IPR036860 SH2_dom_sf
IPR001839 TGF-b_C
IPR029034 Cystine-knot_cytokine
IPR017948 TGFb_CS
IPR015615 TGF-beta-rel
IPR032821 KAsynt_C_assoc
IPR014031 Ketoacyl_synth_C
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR016039 Thiolase-like
IPR014030 Ketoacyl_synth_N
IPR038717 Tc1-like_DDE_dom
SUPFAM
ProteinModelPortal
PDB
5CR4
E-value=1.18984e-07,
Score=130
Ontologies
Topology
Subcellular location
Cytoplasm
Nucleus
Secreted
Nucleus
Secreted
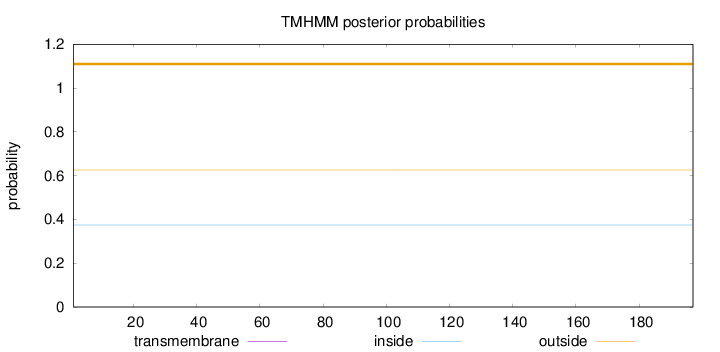
Length:
197
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00063
Exp number, first 60 AAs:
0.00063
Total prob of N-in:
0.37403
outside
1 - 197
Population Genetic Test Statistics
Pi
140.835052
Theta
86.02075
Tajima's D
2.582653
CLR
0.474186
CSRT
0.950302484875756
Interpretation
Uncertain
