Gene
KWMTBOMO14549 Validated by peptides from experiments
Annotation
PREDICTED:_heat_shock_70_kDa_protein_cognate_5-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.461
Sequence
CDS
ATGTTGAACGCGACGCGGGCATTCGGCCGAAAGGCTTTAGAGTGCACCGGATTAAGTTCAGACTTGTATACGCAGAGAAATTTTTCGTCAATTTTGAAAAGCAACGCAACACCCACAGTTCCTATTTACCAGCGACATGGCGTACAATTTAGAAACAAGTCTGAGGGTGTTCGTGGAGCCGTCATTGGAATCGATCTGGGCACGACAAACTCGTGCGTTGCCGTCATGGAGGGCAAGACACCCAAGGTGGTGGAAAACAGTGAAGGGTCTCGGACGACTCCCTCCCATGTGGCCTTCTCGAAAGAGGGCGAGCGTCTGGTCGGAATGCCCGCCAAACGGCAAGCGGTCACGAATAGCGGTAACACGTTTTACGCGACGAAGAGATTGATCGGACGTCGATTCGACGATCCCGAAGTGCAAAAGGACATGAAGAATTTGTCATACAAAGTAGTCAGGGCTTCGAATGGCGACGCCTGGGTGCAAGGTACTGATGGCAAAGTATACTCTCCGAGCCAGATTGGTGCATTTGTGTTGATAAAGATGAAGGAAACTGCTGAAGCATACTTGAACACAAGTGTGAAAAATGCAGTCATAACAGTGCCAGCTTACTTCAATGATTCCCAGCGTCAGGCCACAAAAGATGCAGGTCAAATCTCTGGTTTAAATGTGCTACGAGTTATCAATGAGCCGACTGCTGCTGCCCTCGCTTACGGTATGGACAAGACCGACGACAAAATAATAGCGGTGTATGACCTCGGAGGTGGAACCTTCGATATCTCGGTGCTGGAGATCCAGAAGGGAGTGTTCGAAGTGAAGTCTACCAATGGAGACACTCTGCTGGGAGGTGAAGACTTTGACAATGTTATTGTGAATTTCCTAGTGGATGAGTTTAAACGTGATCAAGGCATTGACATCCGTAAAGACGCTATGGCTATGCAAAGGCTGAAGGAAGCTGCTGAAAAAGCCAAAATTGAATTATCAGGGTCGGTGCAGACTGACATCAACTTGCCTTACCTGACTATGGATGCATCGGGACCCAAACACATGAACTTGAAGATGACACGCTCGAAACTAGAATCTCTAGTTGGGGATCTGATCAGGCGGACGGTGGCGCCGTGCCAGAAGGCCCTTCAGGATGCCGAAGTGCAGCGATCGGACATCGGTGAAGTTCTGCTCGTTGGTGGAATGACGAGAATGCCTAAGGTCCAACAAACAGTACAGGAAATATTCGGCAGGGCTCCGTCTCGAGCCGTGAACCCCGACGAGGCCGTGGCGGTGGGAGCCGCCGTCCAGGGCGGAGTCCTAGCGGGTGACGTCACTGACATCCTTCTCCTCGACGTGACTCCGCTGTCACTGGGCATCGAAACCCTCGGAGGCGTCTTCACCAAGCTGATCACAAGGAACACCACCATACCCACTAAGAAGAGCCAAGTGTTCTCTACCGCGGCCGACGGGCAGACGCAAGTGGAGATCAAGGTCCACCAGGGCGAGCGCGAGATGGCGTCGGACAACAAGCTGCTGGGGCAGTTCTCGCTGGTGGGCATCCCGCCCGCGCCGCGCGGCGTGCCGCAGATAGAGGTCGCCTTCGACATCGACGCCAACGGCATCGTGCACGTGTCCGCGCGGGACAAGGGCACCGGCAAGGAGCAGCAGATCGTGATCCAGTCGTCGGGAGGGCTGTCGAAGGACGAGATCGAGAACATGGTGAAGGCCGCCGAGCAGTTCGCCGCCGCCGACAAGACGCGGCGCGAGCGGGTGGAGGTGGCCAACCAGGCCGAGGCCGTGCTGCACGACACCGACACCAAGATGGACGAGTACAAGGCGCAGCTGCCGCAGGACGAGTGTGACAAACTACGCGAGGAGATGAACAAACTCCGTGAGCTGCTCGCTAAAAAGGACACCATTGAGCCGGAAGAACTTAGAACAGCCACCGGACAGCTGCAGCAGGCCAGCCTTAAACTGTTTGAACAGGCTTACAAGAAGATGGCAGCTGAACGCGAAGGAAGCCAACAACAGCAACAGCAGCAGACGGATCAGACGCAGAGCCAAGAGAAGAAGGAGGAGAAGAATTAG
Protein
MLNATRAFGRKALECTGLSSDLYTQRNFSSILKSNATPTVPIYQRHGVQFRNKSEGVRGAVIGIDLGTTNSCVAVMEGKTPKVVENSEGSRTTPSHVAFSKEGERLVGMPAKRQAVTNSGNTFYATKRLIGRRFDDPEVQKDMKNLSYKVVRASNGDAWVQGTDGKVYSPSQIGAFVLIKMKETAEAYLNTSVKNAVITVPAYFNDSQRQATKDAGQISGLNVLRVINEPTAAALAYGMDKTDDKIIAVYDLGGGTFDISVLEIQKGVFEVKSTNGDTLLGGEDFDNVIVNFLVDEFKRDQGIDIRKDAMAMQRLKEAAEKAKIELSGSVQTDINLPYLTMDASGPKHMNLKMTRSKLESLVGDLIRRTVAPCQKALQDAEVQRSDIGEVLLVGGMTRMPKVQQTVQEIFGRAPSRAVNPDEAVAVGAAVQGGVLAGDVTDILLLDVTPLSLGIETLGGVFTKLITRNTTIPTKKSQVFSTAADGQTQVEIKVHQGEREMASDNKLLGQFSLVGIPPAPRGVPQIEVAFDIDANGIVHVSARDKGTGKEQQIVIQSSGGLSKDEIENMVKAAEQFAAADKTRRERVEVANQAEAVLHDTDTKMDEYKAQLPQDECDKLREEMNKLRELLAKKDTIEPEELRTATGQLQQASLKLFEQAYKKMAAEREGSQQQQQQQTDQTQSQEKKEEKN
Summary
Similarity
Belongs to the heat shock protein 70 family.
Uniprot
I6XKQ0
A0A2W1BDN4
A0A2A4JWZ4
C0KJJ4
S4PBD2
A0A2Z4NA26
+ More
A0A212F5Q1 R4HKY3 A0A0K8TS32 A0A1B0CFI0 A0A1L8DU88 W8GS88 A0A1J1IGQ1 V9IK10 A0A087ZTY7 A0A154PLJ1 D6WA11 A0A195BRI9 A0A158NYJ8 A0A232ER29 A0A2A3EJF9 A0A195FHF1 A0A1B6CN01 A0A1B6I9E9 A0A310SIG6 A0A195DD81 A0A0C9QAV3 A0A2J7QQY0 A0A1S4EGT6 V5TGS6 A0A2P8XLG1 A0A067QIW0 A0A1S5VZG4 A0A1B6LV37 U5EV62 A0A1S5VZC4 A0A0L7RHN2 A0A0J7KVR3 A0A026WAN2 E2B9C9 A0A023EXJ1 A0A0P6IUV3 A0A1Y1M9S6 E0VPP0 A0A2L0V7Q5 A0A182H3F2 A0A182RYW9 A0A034WNJ6 A0A182J031 A0A182TK54 W8BQF5 A0A084VSJ4 A0A182HB17 U4U4E7 A0A151WKA3 N6SVE8 T1DE10 T1D3U9 A0A2R7WEP2 K7JAU7 A0A0K8VZJ8 A0A1Q3F8T0 A0A1Q3F967 A0A1Q3F922 B0WBF6 A0A182VLS8 A0A182YKM1 A0A182WFW9 A0A182PJB1 A0A0A1WWP6 B4KSK2 A0A1S4H618 A0A182X411 A0A182I5I0 B4J6Z0 A0A2M3YYZ2 A0A182FKC1 A0A182NC91 A0A182KCB2 A0A1A9XPQ9 A0A1B0AM43 B4LMH0 A0A2M3YYY1 A0A2M4BG77 A0A182MGM1 Q7PFH8 D3TRQ0 A0A1A9V7P9 A0A182QMZ6 A0A1A9WIA9 T1PA42 A0A2M4A0D2 A0A0P5EIX2 W5JM27 A0A0M4EVJ7 A0A1I8NZY6 B3MDY5 A0A1A9ZPB6 A0A0P5XX42 A0A0A9X596
A0A212F5Q1 R4HKY3 A0A0K8TS32 A0A1B0CFI0 A0A1L8DU88 W8GS88 A0A1J1IGQ1 V9IK10 A0A087ZTY7 A0A154PLJ1 D6WA11 A0A195BRI9 A0A158NYJ8 A0A232ER29 A0A2A3EJF9 A0A195FHF1 A0A1B6CN01 A0A1B6I9E9 A0A310SIG6 A0A195DD81 A0A0C9QAV3 A0A2J7QQY0 A0A1S4EGT6 V5TGS6 A0A2P8XLG1 A0A067QIW0 A0A1S5VZG4 A0A1B6LV37 U5EV62 A0A1S5VZC4 A0A0L7RHN2 A0A0J7KVR3 A0A026WAN2 E2B9C9 A0A023EXJ1 A0A0P6IUV3 A0A1Y1M9S6 E0VPP0 A0A2L0V7Q5 A0A182H3F2 A0A182RYW9 A0A034WNJ6 A0A182J031 A0A182TK54 W8BQF5 A0A084VSJ4 A0A182HB17 U4U4E7 A0A151WKA3 N6SVE8 T1DE10 T1D3U9 A0A2R7WEP2 K7JAU7 A0A0K8VZJ8 A0A1Q3F8T0 A0A1Q3F967 A0A1Q3F922 B0WBF6 A0A182VLS8 A0A182YKM1 A0A182WFW9 A0A182PJB1 A0A0A1WWP6 B4KSK2 A0A1S4H618 A0A182X411 A0A182I5I0 B4J6Z0 A0A2M3YYZ2 A0A182FKC1 A0A182NC91 A0A182KCB2 A0A1A9XPQ9 A0A1B0AM43 B4LMH0 A0A2M3YYY1 A0A2M4BG77 A0A182MGM1 Q7PFH8 D3TRQ0 A0A1A9V7P9 A0A182QMZ6 A0A1A9WIA9 T1PA42 A0A2M4A0D2 A0A0P5EIX2 W5JM27 A0A0M4EVJ7 A0A1I8NZY6 B3MDY5 A0A1A9ZPB6 A0A0P5XX42 A0A0A9X596
Pubmed
28756777
21362495
23622113
22118469
26369729
18362917
+ More
19820115 21347285 28648823 29403074 24845553 28187833 24508170 20798317 24945155 26999592 28004739 20566863 26483478 25348373 24495485 24438588 23537049 24330624 20075255 25244985 25830018 17994087 12364791 20353571 25315136 20920257 23761445 25401762 26823975
19820115 21347285 28648823 29403074 24845553 28187833 24508170 20798317 24945155 26999592 28004739 20566863 26483478 25348373 24495485 24438588 23537049 24330624 20075255 25244985 25830018 17994087 12364791 20353571 25315136 20920257 23761445 25401762 26823975
EMBL
JQ846451
AFN52772.1
KZ150319
PZC71427.1
NWSH01000512
PCG75930.1
+ More
FJ609645 ACM78945.1 GAIX01004676 JAA87884.1 MG745402 AWX67686.1 AGBW02010147 OWR49071.1 HQ647013 AET10425.1 GDAI01000444 JAI17159.1 AJWK01010107 GFDF01004076 JAV10008.1 KF986612 AHK26790.1 CVRI01000048 CRK98946.1 JR051327 AEY61478.1 KQ434968 KZC12703.1 KQ971312 EEZ98080.1 KQ976423 KYM88834.1 ADTU01000682 NNAY01002689 OXU20767.1 KZ288227 PBC31838.1 KQ981606 KYN39434.1 GEDC01022525 JAS14773.1 GECU01024141 JAS83565.1 KQ759849 OAD62568.1 KQ980979 KYN10843.1 GBYB01000339 JAG70106.1 NEVH01012082 PNF30986.1 KF660251 AHB63834.1 PYGN01001782 PSN32841.1 KK853313 KDR08641.1 KX976477 AQP31367.1 GEBQ01012425 JAT27552.1 GANO01002005 JAB57866.1 KX695164 AQP31334.1 KQ414588 KOC70497.1 LBMM01002796 KMQ94354.1 KK107321 EZA52726.1 GL446499 EFN87680.1 GAPW01000424 JAC13174.1 GDUN01000365 JAN95554.1 GEZM01036901 JAV82351.1 DS235366 EEB15346.1 KY914547 MF377632 AVA07270.1 AVM40931.1 JXUM01107366 JXUM01107367 JXUM01107368 JXUM01107369 KQ565135 KXJ71299.1 GAKP01002758 GAKP01002757 JAC56194.1 GAMC01014785 GAMC01014783 GAMC01014781 JAB91774.1 ATLV01016042 KE525054 KFB40938.1 JXUM01123892 KQ566767 KXJ69841.1 KB631924 ERL87223.1 KQ983012 KYQ48288.1 APGK01055062 KB741259 ENN71699.1 GALA01001232 JAA93620.1 GALA01001233 JAA93619.1 KK854718 PTY18133.1 AAZX01008610 GDHF01008055 JAI44259.1 GFDL01011147 JAV23898.1 GFDL01010978 JAV24067.1 GFDL01010970 JAV24075.1 DS231879 EDS42395.1 GBXI01010803 JAD03489.1 CH933808 EDW09507.1 AAAB01008823 APCN01003445 CH916367 EDW02071.1 GGFM01000732 MBW21483.1 JXJN01000335 CH940648 EDW62065.1 GGFM01000728 MBW21479.1 GGFJ01002901 MBW52042.1 AXCM01005795 EAA45310.4 CCAG010018973 EZ424102 ADD20378.1 AXCN02001762 KA645005 AFP59634.1 GGFK01000884 MBW34205.1 GDIP01145842 JAJ77560.1 ADMH02001144 ETN63945.1 CP012524 ALC41657.1 CH902619 EDV37530.1 GDIP01067444 GDIQ01036912 JAM36271.1 JAN57825.1 GBHO01029626 GBHO01029625 GBRD01013078 GBRD01005783 GDHC01014476 JAG13978.1 JAG13979.1 JAG52748.1 JAQ04153.1
FJ609645 ACM78945.1 GAIX01004676 JAA87884.1 MG745402 AWX67686.1 AGBW02010147 OWR49071.1 HQ647013 AET10425.1 GDAI01000444 JAI17159.1 AJWK01010107 GFDF01004076 JAV10008.1 KF986612 AHK26790.1 CVRI01000048 CRK98946.1 JR051327 AEY61478.1 KQ434968 KZC12703.1 KQ971312 EEZ98080.1 KQ976423 KYM88834.1 ADTU01000682 NNAY01002689 OXU20767.1 KZ288227 PBC31838.1 KQ981606 KYN39434.1 GEDC01022525 JAS14773.1 GECU01024141 JAS83565.1 KQ759849 OAD62568.1 KQ980979 KYN10843.1 GBYB01000339 JAG70106.1 NEVH01012082 PNF30986.1 KF660251 AHB63834.1 PYGN01001782 PSN32841.1 KK853313 KDR08641.1 KX976477 AQP31367.1 GEBQ01012425 JAT27552.1 GANO01002005 JAB57866.1 KX695164 AQP31334.1 KQ414588 KOC70497.1 LBMM01002796 KMQ94354.1 KK107321 EZA52726.1 GL446499 EFN87680.1 GAPW01000424 JAC13174.1 GDUN01000365 JAN95554.1 GEZM01036901 JAV82351.1 DS235366 EEB15346.1 KY914547 MF377632 AVA07270.1 AVM40931.1 JXUM01107366 JXUM01107367 JXUM01107368 JXUM01107369 KQ565135 KXJ71299.1 GAKP01002758 GAKP01002757 JAC56194.1 GAMC01014785 GAMC01014783 GAMC01014781 JAB91774.1 ATLV01016042 KE525054 KFB40938.1 JXUM01123892 KQ566767 KXJ69841.1 KB631924 ERL87223.1 KQ983012 KYQ48288.1 APGK01055062 KB741259 ENN71699.1 GALA01001232 JAA93620.1 GALA01001233 JAA93619.1 KK854718 PTY18133.1 AAZX01008610 GDHF01008055 JAI44259.1 GFDL01011147 JAV23898.1 GFDL01010978 JAV24067.1 GFDL01010970 JAV24075.1 DS231879 EDS42395.1 GBXI01010803 JAD03489.1 CH933808 EDW09507.1 AAAB01008823 APCN01003445 CH916367 EDW02071.1 GGFM01000732 MBW21483.1 JXJN01000335 CH940648 EDW62065.1 GGFM01000728 MBW21479.1 GGFJ01002901 MBW52042.1 AXCM01005795 EAA45310.4 CCAG010018973 EZ424102 ADD20378.1 AXCN02001762 KA645005 AFP59634.1 GGFK01000884 MBW34205.1 GDIP01145842 JAJ77560.1 ADMH02001144 ETN63945.1 CP012524 ALC41657.1 CH902619 EDV37530.1 GDIP01067444 GDIQ01036912 JAM36271.1 JAN57825.1 GBHO01029626 GBHO01029625 GBRD01013078 GBRD01005783 GDHC01014476 JAG13978.1 JAG13979.1 JAG52748.1 JAQ04153.1
Proteomes
UP000218220
UP000007151
UP000092461
UP000183832
UP000005203
UP000076502
+ More
UP000007266 UP000078540 UP000005205 UP000215335 UP000242457 UP000078541 UP000078492 UP000235965 UP000079169 UP000245037 UP000027135 UP000053825 UP000036403 UP000053097 UP000008237 UP000009046 UP000069940 UP000249989 UP000075900 UP000075880 UP000075902 UP000030765 UP000030742 UP000075809 UP000019118 UP000002358 UP000002320 UP000075903 UP000076408 UP000075920 UP000075885 UP000009192 UP000076407 UP000075840 UP000001070 UP000069272 UP000075884 UP000075881 UP000092443 UP000092460 UP000008792 UP000075883 UP000007062 UP000092444 UP000078200 UP000075886 UP000091820 UP000095301 UP000000673 UP000092553 UP000095300 UP000007801 UP000092445
UP000007266 UP000078540 UP000005205 UP000215335 UP000242457 UP000078541 UP000078492 UP000235965 UP000079169 UP000245037 UP000027135 UP000053825 UP000036403 UP000053097 UP000008237 UP000009046 UP000069940 UP000249989 UP000075900 UP000075880 UP000075902 UP000030765 UP000030742 UP000075809 UP000019118 UP000002358 UP000002320 UP000075903 UP000076408 UP000075920 UP000075885 UP000009192 UP000076407 UP000075840 UP000001070 UP000069272 UP000075884 UP000075881 UP000092443 UP000092460 UP000008792 UP000075883 UP000007062 UP000092444 UP000078200 UP000075886 UP000091820 UP000095301 UP000000673 UP000092553 UP000095300 UP000007801 UP000092445
PRIDE
Interpro
Gene 3D
ProteinModelPortal
I6XKQ0
A0A2W1BDN4
A0A2A4JWZ4
C0KJJ4
S4PBD2
A0A2Z4NA26
+ More
A0A212F5Q1 R4HKY3 A0A0K8TS32 A0A1B0CFI0 A0A1L8DU88 W8GS88 A0A1J1IGQ1 V9IK10 A0A087ZTY7 A0A154PLJ1 D6WA11 A0A195BRI9 A0A158NYJ8 A0A232ER29 A0A2A3EJF9 A0A195FHF1 A0A1B6CN01 A0A1B6I9E9 A0A310SIG6 A0A195DD81 A0A0C9QAV3 A0A2J7QQY0 A0A1S4EGT6 V5TGS6 A0A2P8XLG1 A0A067QIW0 A0A1S5VZG4 A0A1B6LV37 U5EV62 A0A1S5VZC4 A0A0L7RHN2 A0A0J7KVR3 A0A026WAN2 E2B9C9 A0A023EXJ1 A0A0P6IUV3 A0A1Y1M9S6 E0VPP0 A0A2L0V7Q5 A0A182H3F2 A0A182RYW9 A0A034WNJ6 A0A182J031 A0A182TK54 W8BQF5 A0A084VSJ4 A0A182HB17 U4U4E7 A0A151WKA3 N6SVE8 T1DE10 T1D3U9 A0A2R7WEP2 K7JAU7 A0A0K8VZJ8 A0A1Q3F8T0 A0A1Q3F967 A0A1Q3F922 B0WBF6 A0A182VLS8 A0A182YKM1 A0A182WFW9 A0A182PJB1 A0A0A1WWP6 B4KSK2 A0A1S4H618 A0A182X411 A0A182I5I0 B4J6Z0 A0A2M3YYZ2 A0A182FKC1 A0A182NC91 A0A182KCB2 A0A1A9XPQ9 A0A1B0AM43 B4LMH0 A0A2M3YYY1 A0A2M4BG77 A0A182MGM1 Q7PFH8 D3TRQ0 A0A1A9V7P9 A0A182QMZ6 A0A1A9WIA9 T1PA42 A0A2M4A0D2 A0A0P5EIX2 W5JM27 A0A0M4EVJ7 A0A1I8NZY6 B3MDY5 A0A1A9ZPB6 A0A0P5XX42 A0A0A9X596
A0A212F5Q1 R4HKY3 A0A0K8TS32 A0A1B0CFI0 A0A1L8DU88 W8GS88 A0A1J1IGQ1 V9IK10 A0A087ZTY7 A0A154PLJ1 D6WA11 A0A195BRI9 A0A158NYJ8 A0A232ER29 A0A2A3EJF9 A0A195FHF1 A0A1B6CN01 A0A1B6I9E9 A0A310SIG6 A0A195DD81 A0A0C9QAV3 A0A2J7QQY0 A0A1S4EGT6 V5TGS6 A0A2P8XLG1 A0A067QIW0 A0A1S5VZG4 A0A1B6LV37 U5EV62 A0A1S5VZC4 A0A0L7RHN2 A0A0J7KVR3 A0A026WAN2 E2B9C9 A0A023EXJ1 A0A0P6IUV3 A0A1Y1M9S6 E0VPP0 A0A2L0V7Q5 A0A182H3F2 A0A182RYW9 A0A034WNJ6 A0A182J031 A0A182TK54 W8BQF5 A0A084VSJ4 A0A182HB17 U4U4E7 A0A151WKA3 N6SVE8 T1DE10 T1D3U9 A0A2R7WEP2 K7JAU7 A0A0K8VZJ8 A0A1Q3F8T0 A0A1Q3F967 A0A1Q3F922 B0WBF6 A0A182VLS8 A0A182YKM1 A0A182WFW9 A0A182PJB1 A0A0A1WWP6 B4KSK2 A0A1S4H618 A0A182X411 A0A182I5I0 B4J6Z0 A0A2M3YYZ2 A0A182FKC1 A0A182NC91 A0A182KCB2 A0A1A9XPQ9 A0A1B0AM43 B4LMH0 A0A2M3YYY1 A0A2M4BG77 A0A182MGM1 Q7PFH8 D3TRQ0 A0A1A9V7P9 A0A182QMZ6 A0A1A9WIA9 T1PA42 A0A2M4A0D2 A0A0P5EIX2 W5JM27 A0A0M4EVJ7 A0A1I8NZY6 B3MDY5 A0A1A9ZPB6 A0A0P5XX42 A0A0A9X596
PDB
2KHO
E-value=0,
Score=1679
Ontologies
GO
PANTHER
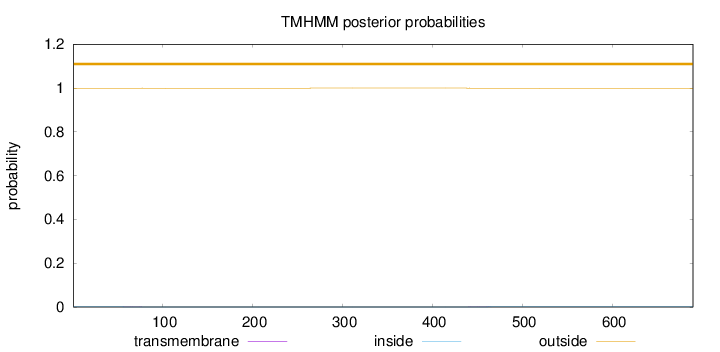
Topology
Length:
690
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02371
Exp number, first 60 AAs:
0.00145
Total prob of N-in:
0.00076
outside
1 - 690
Population Genetic Test Statistics
Pi
252.27513
Theta
206.427178
Tajima's D
1.121877
CLR
0.244477
CSRT
0.692465376731163
Interpretation
Uncertain
