Gene
KWMTBOMO14546 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014403
Annotation
PREDICTED:_uncharacterized_protein_LOC106115633_isoform_X1_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.858 PlasmaMembrane Reliability : 1.893
Sequence
CDS
ATGGCGTTTTGCCGTTTACTGTTCCCGTTGCTAGTTCTAAGCGTGGCCAGTGCGTCGACGGTGCCAGTGTTCTTATGGGGCGACTTATCCAGTAGCTCTGGAAACTCCCTGAAATCGAACCCATTGAGCACGGTGTCCGCTCAAGGGTTCAGCGAAATTTTGAATAATGAGCTGAAAGACGACCCCTACGTGATAGTATTCCTTGAAGAAACCTTATCGGTCGAAGATTTTTCACGTAAAAATAACGAAGGTGATATTTCGATTCCGTACCTGTATGATAATTTAAGTGACGCTCTGTACTTGCCTTCCGTCGAAGATGCCTCAAGAGTACTCGAGGAGGTCGCCAAAGGAGCTGTTCATGTAAAACTCACCCAAAACGGACTCTCTGCAGCCATTGAAGATAAAAATAACAGGTTTATGTTCATCGCCTTGAAGGATGCTATGGAAGGCGAGTCACGTTACGACCTCCTCCGGCGCCATAGCAACTTTATGGAGGACCAGATATCTAAGCTGGAGAAGAAAGGTGATGTAATAGCTGTATACACTGCCCGAAACCCATCATGGACCATTCCGGACACCCACTCAAGGGTTCGTCGTCAAGCCAACTCTTCTGTAACCAACGGACCTGATTACTCATTAGACGGTCTGCGTTTGTACTTAACCGGCGTTACATTAGACCACAGCAATGCCAGCGTGACATTCACAAACTATGCAGGCGGCTCATCTGAATACAATACAACAACTGGAGTCTTGAACGGCACTTTAAACTTTGATCAGGGAAGCCTGACTCTTAACTTCCACAGGGCCGCAGGATATTGGTTCTTTGACACCGTGCACTTTGTCCAGACCAATCCGGCTGTTGACGAGATCCTCTATGCCACTGACGACTTGAATGCCCTGCTCGACTTTTCTTATCGTTGCAAACAGTCTGTCAGCTTCAAGAGTATCAACGAGACACAAGAGTACAAGGTGTCGTTTATCGATATAAAGGTGCAGCCGTTCTTTGGTCCAACAAACGCTTCGACTCCGTTTGGGGATTCCTTTAACTGCGTCGGATTCTTCTCTGCCCCCATCTGGTCCGGTCTGTTCGTTGTGTTCATATTGCTGTCCATAACCTTCTATGGCATCATGACCATGATGGACATTAAAACCATGGATAGGTTCGATGACCCGAAAGGCAAAACCATTACCATAAACGCAGGAGAGTAA
Protein
MAFCRLLFPLLVLSVASASTVPVFLWGDLSSSSGNSLKSNPLSTVSAQGFSEILNNELKDDPYVIVFLEETLSVEDFSRKNNEGDISIPYLYDNLSDALYLPSVEDASRVLEEVAKGAVHVKLTQNGLSAAIEDKNNRFMFIALKDAMEGESRYDLLRRHSNFMEDQISKLEKKGDVIAVYTARNPSWTIPDTHSRVRRQANSSVTNGPDYSLDGLRLYLTGVTLDHSNASVTFTNYAGGSSEYNTTTGVLNGTLNFDQGSLTLNFHRAAGYWFFDTVHFVQTNPAVDEILYATDDLNALLDFSYRCKQSVSFKSINETQEYKVSFIDIKVQPFFGPTNASTPFGDSFNCVGFFSAPIWSGLFVVFILLSITFYGIMTMMDIKTMDRFDDPKGKTITINAGE
Summary
Uniprot
H9JXY5
A0A212ESJ8
A0A1E1WHE2
A0A0N1IPQ5
A0A2A4JX12
A0A2H1V8X3
+ More
A0A194Q0J9 A0A2W1B5E4 A0A3S2LA54 A0A0N1IMP9 A0A0L7LJ85 V5GYH4 A0A067QVB5 A0A2J7R414 A0A1Y1NDP6 D6W7U7 A0A1W4XGF7 A0A067R8J1 A0A2P8YCK1 A0A1B6MNA2 B3N7J2 A0A2J7R2V1 A0A1I8NRR8 A0A2P8YR64 T1PB59 A0A224XKN8 A0A182LAG0 Q7QFR7 A0A182I8K6 A0A182X1P9 A0A182TWT6 B4LJ75 A0A182VL06 Q7JR49 Q28ZI1 Q1HRK0 B4P3Z4 A0A1W4UZU2 B4GHQ8 E0VV03 B4KPN7 A0A1B6DFB7 A0A182QEN0 B4J7Q3 A0A182VQB1 A0A182P5S7 A0A023ER79 A0A182N6Y0 A0A182RBU0 A0A182GMH7 A0A1L8EEU0 A0A1L8EEP1 A0A1L8EEW2 A0A1Z5KW60 T1HAB1 A0A182MMC6 B3MFM0 A0A1L8EEY3 A0A182F9E0 A0A0M5JAB9 A0A2A3EQU0 A0A1J1I9A9 A0A182K4B0 A0A2M4A0D4 A0A088AF62 T1DEB0 W8BV42 A0A310S6L0 A0A3B0JP19 A0A182J280 A0A2M4BQC2 A0A2M3YYS2 A0A2M4BQC6 B4QH65 B4N5Z4 A0A182SES8 A0A2R7VYQ3 B4HSP7 T1E9T9 U5EP38 A0A084VDR9 A0A0A1WSY1 E2A4R1 A0A0K8WGH8 A0A034VLY5 A0A0K8UGG1 A0A0K8UMA7 A0A0T6B0Z3 W5J4K7 A0A182XX75 A0A154P1V0 C4WW04 A0A0C9QH31 T1IUV9
A0A194Q0J9 A0A2W1B5E4 A0A3S2LA54 A0A0N1IMP9 A0A0L7LJ85 V5GYH4 A0A067QVB5 A0A2J7R414 A0A1Y1NDP6 D6W7U7 A0A1W4XGF7 A0A067R8J1 A0A2P8YCK1 A0A1B6MNA2 B3N7J2 A0A2J7R2V1 A0A1I8NRR8 A0A2P8YR64 T1PB59 A0A224XKN8 A0A182LAG0 Q7QFR7 A0A182I8K6 A0A182X1P9 A0A182TWT6 B4LJ75 A0A182VL06 Q7JR49 Q28ZI1 Q1HRK0 B4P3Z4 A0A1W4UZU2 B4GHQ8 E0VV03 B4KPN7 A0A1B6DFB7 A0A182QEN0 B4J7Q3 A0A182VQB1 A0A182P5S7 A0A023ER79 A0A182N6Y0 A0A182RBU0 A0A182GMH7 A0A1L8EEU0 A0A1L8EEP1 A0A1L8EEW2 A0A1Z5KW60 T1HAB1 A0A182MMC6 B3MFM0 A0A1L8EEY3 A0A182F9E0 A0A0M5JAB9 A0A2A3EQU0 A0A1J1I9A9 A0A182K4B0 A0A2M4A0D4 A0A088AF62 T1DEB0 W8BV42 A0A310S6L0 A0A3B0JP19 A0A182J280 A0A2M4BQC2 A0A2M3YYS2 A0A2M4BQC6 B4QH65 B4N5Z4 A0A182SES8 A0A2R7VYQ3 B4HSP7 T1E9T9 U5EP38 A0A084VDR9 A0A0A1WSY1 E2A4R1 A0A0K8WGH8 A0A034VLY5 A0A0K8UGG1 A0A0K8UMA7 A0A0T6B0Z3 W5J4K7 A0A182XX75 A0A154P1V0 C4WW04 A0A0C9QH31 T1IUV9
Pubmed
19121390
22118469
26354079
28756777
26227816
24845553
+ More
28004739 18362917 19820115 29403074 17994087 25315136 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23185243 17204158 17510324 17550304 20566863 24945155 26483478 28528879 24330624 24495485 22936249 24438588 25830018 20798317 25348373 20920257 23761445 25244985
28004739 18362917 19820115 29403074 17994087 25315136 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23185243 17204158 17510324 17550304 20566863 24945155 26483478 28528879 24330624 24495485 22936249 24438588 25830018 20798317 25348373 20920257 23761445 25244985
EMBL
BABH01041376
AGBW02012775
OWR44472.1
GDQN01004728
JAT86326.1
KQ460130
+ More
KPJ17529.1 NWSH01000512 PCG75932.1 ODYU01001279 SOQ37278.1 KQ459581 KPI99107.1 KZ150319 PZC71429.1 RSAL01000072 RVE49084.1 KQ459574 KPI99574.1 JTDY01000973 KOB75241.1 GALX01003018 JAB65448.1 KK852908 KDR13979.1 NEVH01007816 PNF35574.1 GEZM01006604 JAV95688.1 KQ971307 EFA11226.1 KK852669 KDR18876.1 PYGN01000704 PSN41978.1 GEBQ01002540 JAT37437.1 CH954177 EDV59397.1 NEVH01007825 PNF35171.1 PYGN01000418 PSN46733.1 KA645208 AFP59837.1 GFTR01006038 JAW10388.1 AAAB01008844 EAA06051.5 APCN01003080 CH940648 EDW61511.1 AE013599 BT003279 AAF58965.1 AAF58966.2 AAO25036.1 CM000071 EAL25632.1 KRT02422.1 DQ440094 CH477465 ABF18127.1 EAT40488.1 CM000157 EDW89477.1 CH479183 EDW36028.1 DS235797 EEB17209.1 CH933808 EDW09147.1 GEDC01012914 JAS24384.1 AXCN02000314 CH916367 EDW02201.1 GAPW01001945 JAC11653.1 JXUM01074646 KQ562843 KXJ75025.1 GFDG01001603 JAV17196.1 GFDG01001602 JAV17197.1 GFDG01001605 JAV17194.1 GFJQ02007651 JAV99318.1 ACPB03016232 AXCM01008102 CH902619 EDV36705.2 GFDG01001604 JAV17195.1 CP012524 ALC42044.1 KZ288199 PBC33632.1 CVRI01000043 CRK96156.1 GGFK01000888 MBW34209.1 GALA01001107 JAA93745.1 GAMC01003508 JAC03048.1 KQ767877 OAD53277.1 OUUW01000001 SPP72938.1 GGFJ01006145 MBW55286.1 GGFM01000656 MBW21407.1 GGFJ01006146 MBW55287.1 CM000362 CM002911 EDX06319.1 KMY92481.1 CH964154 EDW79783.1 KK854166 PTY12449.1 CH480816 EDW47074.1 GAMD01001017 JAB00574.1 GANO01004895 JAB54976.1 ATLV01011716 KE524713 KFB36113.1 GBXI01012345 JAD01947.1 GL436716 EFN71595.1 GDHF01002076 JAI50238.1 GAKP01016152 JAC42800.1 GDHF01026613 JAI25701.1 GDHF01024613 JAI27701.1 LJIG01016297 KRT81025.1 ADMH02002112 ETN58856.1 KQ434803 KZC05906.1 AK341770 BAH72074.1 GBYB01013953 JAG83720.1 JH431566
KPJ17529.1 NWSH01000512 PCG75932.1 ODYU01001279 SOQ37278.1 KQ459581 KPI99107.1 KZ150319 PZC71429.1 RSAL01000072 RVE49084.1 KQ459574 KPI99574.1 JTDY01000973 KOB75241.1 GALX01003018 JAB65448.1 KK852908 KDR13979.1 NEVH01007816 PNF35574.1 GEZM01006604 JAV95688.1 KQ971307 EFA11226.1 KK852669 KDR18876.1 PYGN01000704 PSN41978.1 GEBQ01002540 JAT37437.1 CH954177 EDV59397.1 NEVH01007825 PNF35171.1 PYGN01000418 PSN46733.1 KA645208 AFP59837.1 GFTR01006038 JAW10388.1 AAAB01008844 EAA06051.5 APCN01003080 CH940648 EDW61511.1 AE013599 BT003279 AAF58965.1 AAF58966.2 AAO25036.1 CM000071 EAL25632.1 KRT02422.1 DQ440094 CH477465 ABF18127.1 EAT40488.1 CM000157 EDW89477.1 CH479183 EDW36028.1 DS235797 EEB17209.1 CH933808 EDW09147.1 GEDC01012914 JAS24384.1 AXCN02000314 CH916367 EDW02201.1 GAPW01001945 JAC11653.1 JXUM01074646 KQ562843 KXJ75025.1 GFDG01001603 JAV17196.1 GFDG01001602 JAV17197.1 GFDG01001605 JAV17194.1 GFJQ02007651 JAV99318.1 ACPB03016232 AXCM01008102 CH902619 EDV36705.2 GFDG01001604 JAV17195.1 CP012524 ALC42044.1 KZ288199 PBC33632.1 CVRI01000043 CRK96156.1 GGFK01000888 MBW34209.1 GALA01001107 JAA93745.1 GAMC01003508 JAC03048.1 KQ767877 OAD53277.1 OUUW01000001 SPP72938.1 GGFJ01006145 MBW55286.1 GGFM01000656 MBW21407.1 GGFJ01006146 MBW55287.1 CM000362 CM002911 EDX06319.1 KMY92481.1 CH964154 EDW79783.1 KK854166 PTY12449.1 CH480816 EDW47074.1 GAMD01001017 JAB00574.1 GANO01004895 JAB54976.1 ATLV01011716 KE524713 KFB36113.1 GBXI01012345 JAD01947.1 GL436716 EFN71595.1 GDHF01002076 JAI50238.1 GAKP01016152 JAC42800.1 GDHF01026613 JAI25701.1 GDHF01024613 JAI27701.1 LJIG01016297 KRT81025.1 ADMH02002112 ETN58856.1 KQ434803 KZC05906.1 AK341770 BAH72074.1 GBYB01013953 JAG83720.1 JH431566
Proteomes
UP000005204
UP000007151
UP000053240
UP000218220
UP000053268
UP000283053
+ More
UP000037510 UP000027135 UP000235965 UP000007266 UP000192223 UP000245037 UP000008711 UP000095300 UP000095301 UP000075882 UP000007062 UP000075840 UP000076407 UP000075902 UP000008792 UP000075903 UP000000803 UP000001819 UP000008820 UP000002282 UP000192221 UP000008744 UP000009046 UP000009192 UP000075886 UP000001070 UP000075920 UP000075885 UP000075884 UP000075900 UP000069940 UP000249989 UP000015103 UP000075883 UP000007801 UP000069272 UP000092553 UP000242457 UP000183832 UP000075881 UP000005203 UP000268350 UP000075880 UP000000304 UP000007798 UP000075901 UP000001292 UP000030765 UP000000311 UP000000673 UP000076408 UP000076502
UP000037510 UP000027135 UP000235965 UP000007266 UP000192223 UP000245037 UP000008711 UP000095300 UP000095301 UP000075882 UP000007062 UP000075840 UP000076407 UP000075902 UP000008792 UP000075903 UP000000803 UP000001819 UP000008820 UP000002282 UP000192221 UP000008744 UP000009046 UP000009192 UP000075886 UP000001070 UP000075920 UP000075885 UP000075884 UP000075900 UP000069940 UP000249989 UP000015103 UP000075883 UP000007801 UP000069272 UP000092553 UP000242457 UP000183832 UP000075881 UP000005203 UP000268350 UP000075880 UP000000304 UP000007798 UP000075901 UP000001292 UP000030765 UP000000311 UP000000673 UP000076408 UP000076502
Pfam
PF05827 ATP-synt_S1
Interpro
IPR008388
ATPase_V1-cplx_s1su
ProteinModelPortal
H9JXY5
A0A212ESJ8
A0A1E1WHE2
A0A0N1IPQ5
A0A2A4JX12
A0A2H1V8X3
+ More
A0A194Q0J9 A0A2W1B5E4 A0A3S2LA54 A0A0N1IMP9 A0A0L7LJ85 V5GYH4 A0A067QVB5 A0A2J7R414 A0A1Y1NDP6 D6W7U7 A0A1W4XGF7 A0A067R8J1 A0A2P8YCK1 A0A1B6MNA2 B3N7J2 A0A2J7R2V1 A0A1I8NRR8 A0A2P8YR64 T1PB59 A0A224XKN8 A0A182LAG0 Q7QFR7 A0A182I8K6 A0A182X1P9 A0A182TWT6 B4LJ75 A0A182VL06 Q7JR49 Q28ZI1 Q1HRK0 B4P3Z4 A0A1W4UZU2 B4GHQ8 E0VV03 B4KPN7 A0A1B6DFB7 A0A182QEN0 B4J7Q3 A0A182VQB1 A0A182P5S7 A0A023ER79 A0A182N6Y0 A0A182RBU0 A0A182GMH7 A0A1L8EEU0 A0A1L8EEP1 A0A1L8EEW2 A0A1Z5KW60 T1HAB1 A0A182MMC6 B3MFM0 A0A1L8EEY3 A0A182F9E0 A0A0M5JAB9 A0A2A3EQU0 A0A1J1I9A9 A0A182K4B0 A0A2M4A0D4 A0A088AF62 T1DEB0 W8BV42 A0A310S6L0 A0A3B0JP19 A0A182J280 A0A2M4BQC2 A0A2M3YYS2 A0A2M4BQC6 B4QH65 B4N5Z4 A0A182SES8 A0A2R7VYQ3 B4HSP7 T1E9T9 U5EP38 A0A084VDR9 A0A0A1WSY1 E2A4R1 A0A0K8WGH8 A0A034VLY5 A0A0K8UGG1 A0A0K8UMA7 A0A0T6B0Z3 W5J4K7 A0A182XX75 A0A154P1V0 C4WW04 A0A0C9QH31 T1IUV9
A0A194Q0J9 A0A2W1B5E4 A0A3S2LA54 A0A0N1IMP9 A0A0L7LJ85 V5GYH4 A0A067QVB5 A0A2J7R414 A0A1Y1NDP6 D6W7U7 A0A1W4XGF7 A0A067R8J1 A0A2P8YCK1 A0A1B6MNA2 B3N7J2 A0A2J7R2V1 A0A1I8NRR8 A0A2P8YR64 T1PB59 A0A224XKN8 A0A182LAG0 Q7QFR7 A0A182I8K6 A0A182X1P9 A0A182TWT6 B4LJ75 A0A182VL06 Q7JR49 Q28ZI1 Q1HRK0 B4P3Z4 A0A1W4UZU2 B4GHQ8 E0VV03 B4KPN7 A0A1B6DFB7 A0A182QEN0 B4J7Q3 A0A182VQB1 A0A182P5S7 A0A023ER79 A0A182N6Y0 A0A182RBU0 A0A182GMH7 A0A1L8EEU0 A0A1L8EEP1 A0A1L8EEW2 A0A1Z5KW60 T1HAB1 A0A182MMC6 B3MFM0 A0A1L8EEY3 A0A182F9E0 A0A0M5JAB9 A0A2A3EQU0 A0A1J1I9A9 A0A182K4B0 A0A2M4A0D4 A0A088AF62 T1DEB0 W8BV42 A0A310S6L0 A0A3B0JP19 A0A182J280 A0A2M4BQC2 A0A2M3YYS2 A0A2M4BQC6 B4QH65 B4N5Z4 A0A182SES8 A0A2R7VYQ3 B4HSP7 T1E9T9 U5EP38 A0A084VDR9 A0A0A1WSY1 E2A4R1 A0A0K8WGH8 A0A034VLY5 A0A0K8UGG1 A0A0K8UMA7 A0A0T6B0Z3 W5J4K7 A0A182XX75 A0A154P1V0 C4WW04 A0A0C9QH31 T1IUV9
Ontologies
KEGG
PATHWAY
PANTHER
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.976165
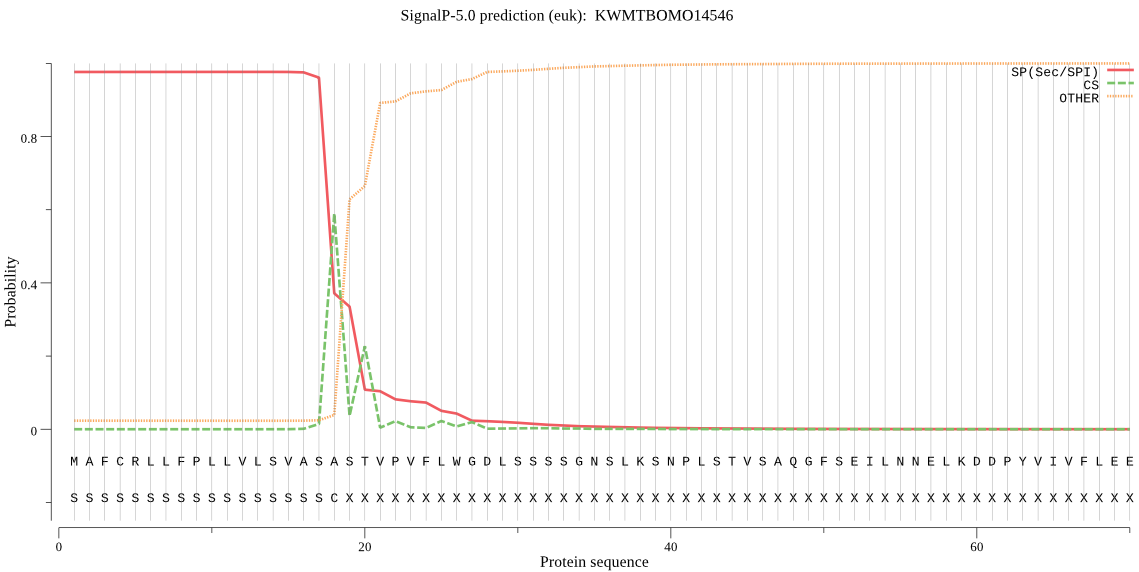
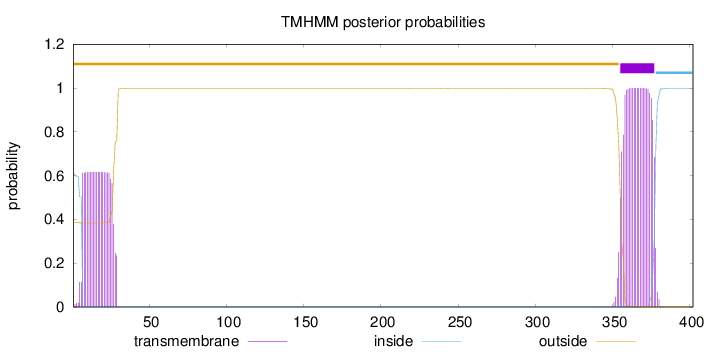
Length:
402
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
35.61112
Exp number, first 60 AAs:
13.36283
Total prob of N-in:
0.61439
POSSIBLE N-term signal
sequence
outside
1 - 354
TMhelix
355 - 377
inside
378 - 402
Population Genetic Test Statistics
Pi
25.969204
Theta
184.223016
Tajima's D
0.870062
CLR
0.254657
CSRT
0.624668766561672
Interpretation
Uncertain
