Gene
KWMTBOMO14536 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014294
Annotation
PREDICTED:_ATP-citrate_synthase_[Amyelois_transitella]
Full name
ATP-citrate synthase
Alternative Name
ATP-citrate (pro-S-)-lyase
Citrate cleavage enzyme
Citrate cleavage enzyme
Location in the cell
Cytoplasmic Reliability : 2.071
Sequence
CDS
ATGTCGTCGAAAGCTATCTACGAGGCAACGGGAAAGGATCTGATCAACAGACACATAGCCTCCGGTACGGCTCTGGTACCGTGCAGGTTCGCCACATTCGAACAGAGCACAGTGTGGGAGGATGAGCTGGGAAAAAATCCATGGCTCTCGAAGGAGCAATTGGTCGTGAAGCCGGACCAGCTAATCAAGCGTCGCGGCAAACTGGGTCTGGTCGGCGTGAACAAGACTGCGGCGGAGGTCCGGCGTTGGCTCGGCGAGCACATGGCCAAGGAGCAGACGATAGGGGCCTGCGTCGGGCGGCTGAGGCACTTTATAGTCGAGCCCTTCGTCAAACACGAACCGAGCGACGAGATGTACCTGTGCATACAGTCGGGCCGGCGCTCCGACACCATCCTGTTCCACCACCAGGGAGGGGTCGACGTCGGAGATGTGGACGCCCTGGCCATCCGACTAGAGGTGCCCGTGGACACCTTCCCGACGGGAGAAGAAGTCGAGAGGGTTTTGTTGAAGAACGTCAAATCTCCATCTACTAAAAGGACCCTGACGACGTTCATAGTGTCTCTATACCGCGTCTACATTGACTTGTACTTCACGTACATGGAGATCAATCCGCTGGTGGTGACCAACGAGAGGATATACCTGCTGGACCTGGCCGCGAAACTGGACCAGACCGCGGACTTCATCTGCGCGAAGAACTGGGGTGAGGTCACGTTCCCTCCGCCCTTCGGGAGGGACGCCTACCCTGAAGAAGCGCACATCGCCGACCTGGACGCCAAGAGCGGGGCCAGTCTGAAGTTGACGGTGCTGAACAAGTCTGGTCGCGTGTGGACGATGGTGGCGGGCGGCGGCGCCAGCGTGGTGTACACGGACACGGTGTGCGCGCTGGGCGGCGCTCACGACCTGGCCAACTACGGGGAGTACTCGGGCGCGCCCACGGAGACGCAGACCGCGGACTACGCGAAGACCATCTTCAGCCTCATGTGCCGCGAGAAGCATCCCGACGGGAAGGTACTGATCGTGGGTGGCGGCATCGCCAACTTCACCAACGTGGCGGATACCTTCCGGGGCATCATCACCGCCATAGAGACGTACAAGGACGCCCTAAAGCAGTACAACGTCACCATCTTCGTGAGGAGAGGGGGACCAAACTACCAGGAGGGACTCAGACAAATGCGCGAAGTGGGCCAGCGCCTCCGCATCCCGCTGTACGTGTTCGGTCCGGAGACGAACATGACGTCCATAGTGGGCCTGGCGCTGGGACACTCGCCCATACCCAAGGAGCACCAGCTCGACTACGCGCCCAAGCAGCTGCCCAGGCCGCAGCCAGCGCCGGCTCCGAAACTGGACCTGCCTGACGTGACCCACAGCCTGCTCGACCTGATCTGCTCCCAGGCTCCGACCAAGCTCGACCTGGGCCAGGGCGTCTCCACCTCGATGCCCCTCTTCAGCGATAGGACGAAGGCCATCATATGGGGCATGCAGAATAGGGCCATACAGGGCATGTTGGACTTCGACTACATCTGCCGGCGCTCGGAGCCCTCGGTCGTCGCCATCGTGTACCCGTTCACGGCGGACCACAAACAGAAGTACTACTTCGGGAACAAGGAAGTCTTCATACCGGTGTTCTCGGACATGGACGTGGCGATGCGCAAGCACCAGGAGGCGACGGTACTCGTCAACTTCGCTTCGCTTCGCTCGGCCTACGACAGCACCATGCAGGCCATGCAGCACCCTCAGATCAACACCGTGGTCATCATCGCGGAGGGCATCCCCGAGAACATGACGCGGAAGATCATCAAGCTCGCTGACGTCAGAGGGGTGAACATAATCGGGCCTGCCACTGTGGGAGGCATTAAACCCGGCTGCTTCAAGATCGGTAACACGGCTGGCATGATTGACAATATCATTGACAGCAAACTGTACCGGCCTGGGAGCGTGGCGTACGTGTCCCGCTCGGGAGGCATGAGCAACGAGCTGAACAAGATCCTGTCGAAGGACGCGGACGGCGTGTGCGAGGGGGTCGCCATCGGGGGCGACCGCTACCCTGGGACCACCTTCATCGACCACCTGCTCAGGTTCGAAGCGGACCCGCAAGTGAAGATGCTGGTGCTGCTTGGAGAAGTGGGGGGAGTCGAGGAGTACCACGTCTGCAAGGCCATCAGGGACGGAATCATCAAGAAGCCCCTCGTGGCCTGGTGCATAGGAACTTGCTCCGACATGTTCACGTCTGAGGTCCAGTTCGGACACGCGGGCTCGCTGGCTGGCTCCGCCGTTGAGAAGGCTGCGGTCAAGAATGAGGCTCTGAAGGCCTACGGAGCGACCGTGCCGGACTCCTTCGACGGCCTAGGGGCTGCCGTGGGCGAGGTCTACCGCAGGCTGGTCGACGAGGGCAAGATCATCGTCAAGGAGGAGGTGGAACCGCCCAAGGTCCCCATGGACTACGATTGGGCTCGGAAGTTGGGTATAATCCGGAAGCCGGCCGCCTTCATCAGCACGATATGCGACGAGCGAGGCCAGGAGCTGAGCTACTGCGGGGTGCCCATCTCCCAGGTGCTGGAGAGGCAGCTCGGGGTCGGCGGCACCGTCAGCCTGCTGTGGTTCCAGCGCGAGCTGCCGGAGTGGGCGAGCCGGTTCTTCGAGCTGGTGCTGATAGTGACCGCGGACCACGGGCCCGCCGTGTCGGGCGCGCACAACACCATGGTCACGGCGCGCGCCGGCAAGGACCTCATCTCCTCCGTCGTCAGCGGCCTGCTCACCATCGGCGACCGGTTCGGTGGGGCCCTGGACCGCGCCGCGGCTGACTTCTGCGCCGCCTACGACAAGGGACAGCACCCCCAGGAGTTCGTCAACGAGAAGAGGGCCAAGGGGGAGCTGATCATGGGCATCGGACACAGGGTGAAGTCGATAAACAACCCGGACTCCCGCGTGCGCGAGCTGAAGGCGTACGTGACGGCGCGCTGGCCCGCGTGGCCGGTCACCCGGTACGCGCTCGACGTGGAGGCCATCACCACCAGGAAGAAGCCGAACCTCATCCTCAACGTGGACGGCATCGTGGCCGCTGCCATGGTCGACCTGTTCCGCCACTGCCAGCTCTTCACGCAAGATGAAGGCAATAATTATATTCAGATGGGCTCAATCAACGCTCTGTTCGTCCTGGGTCGCACGATAGGTCTGGTCGGACATTACTTGGACCAGAAGAGGATGAAGCAGCCTCTGTACCGTCATCCGTGGGACGACATCACCTACATGTCGCCTCTGAACTAA
Protein
MSSKAIYEATGKDLINRHIASGTALVPCRFATFEQSTVWEDELGKNPWLSKEQLVVKPDQLIKRRGKLGLVGVNKTAAEVRRWLGEHMAKEQTIGACVGRLRHFIVEPFVKHEPSDEMYLCIQSGRRSDTILFHHQGGVDVGDVDALAIRLEVPVDTFPTGEEVERVLLKNVKSPSTKRTLTTFIVSLYRVYIDLYFTYMEINPLVVTNERIYLLDLAAKLDQTADFICAKNWGEVTFPPPFGRDAYPEEAHIADLDAKSGASLKLTVLNKSGRVWTMVAGGGASVVYTDTVCALGGAHDLANYGEYSGAPTETQTADYAKTIFSLMCREKHPDGKVLIVGGGIANFTNVADTFRGIITAIETYKDALKQYNVTIFVRRGGPNYQEGLRQMREVGQRLRIPLYVFGPETNMTSIVGLALGHSPIPKEHQLDYAPKQLPRPQPAPAPKLDLPDVTHSLLDLICSQAPTKLDLGQGVSTSMPLFSDRTKAIIWGMQNRAIQGMLDFDYICRRSEPSVVAIVYPFTADHKQKYYFGNKEVFIPVFSDMDVAMRKHQEATVLVNFASLRSAYDSTMQAMQHPQINTVVIIAEGIPENMTRKIIKLADVRGVNIIGPATVGGIKPGCFKIGNTAGMIDNIIDSKLYRPGSVAYVSRSGGMSNELNKILSKDADGVCEGVAIGGDRYPGTTFIDHLLRFEADPQVKMLVLLGEVGGVEEYHVCKAIRDGIIKKPLVAWCIGTCSDMFTSEVQFGHAGSLAGSAVEKAAVKNEALKAYGATVPDSFDGLGAAVGEVYRRLVDEGKIIVKEEVEPPKVPMDYDWARKLGIIRKPAAFISTICDERGQELSYCGVPISQVLERQLGVGGTVSLLWFQRELPEWASRFFELVLIVTADHGPAVSGAHNTMVTARAGKDLISSVVSGLLTIGDRFGGALDRAAADFCAAYDKGQHPQEFVNEKRAKGELIMGIGHRVKSINNPDSRVRELKAYVTARWPAWPVTRYALDVEAITTRKKPNLILNVDGIVAAAMVDLFRHCQLFTQDEGNNYIQMGSINALFVLGRTIGLVGHYLDQKRMKQPLYRHPWDDITYMSPLN
Summary
Description
ATP-citrate synthase is the primary enzyme responsible for the synthesis of cytosolic acetyl-CoA in many tissues.
Catalytic Activity
acetyl-CoA + ADP + oxaloacetate + phosphate = ATP + citrate + CoA
Subunit
Homotetramer.
Similarity
In the C-terminal section; belongs to the succinate/malate CoA ligase alpha subunit family.
In the N-terminal section; belongs to the succinate/malate CoA ligase beta subunit family.
In the N-terminal section; belongs to the succinate/malate CoA ligase beta subunit family.
Uniprot
S4PEX5
A0A0N0P9L9
A0A194R999
A0A1Y1LMG7
A0A1W4WDM4
A0A1W4WEX6
+ More
V5GTA5 D2A5W6 A0A182U8G4 A0A182HQV2 A0A2J7RDY5 W8AYI0 A0A1A9XP97 A0A182VKF6 A0A182XLF6 A0A2J7RDZ4 A0A0A1X3Z2 A0A067QJK7 A0A1B0CWI3 A0A0V0G5Z5 A0A182PCH9 A0A224XIL5 A0A182R7Q4 A0A182WA54 A0A034VWU5 A0A1L8E1A7 A0A182Q238 U5EZ83 A0A2M3ZZ32 A0A1A9W2F7 A0A0J9RDY6 A0A1L8E112 A0A2M3ZZ25 A0A0K8U6N3 B4P6T1 A0A0L0CF01 W5J1A9 A0A2M3YY31 A0A182NPW4 A0A0B4LFH8 A0A1W4UZZ4 A0A1L8E0T3 A0A182JI57 A0A182YIL9 B4QHD4 A0A0M4EUF0 A0A182K9C8 B4HSH9 A0A182MUM6 A0A0R1DSP7 A0A069DXN3 A0A2M4BBA3 E2QCF1 A0A1A9ZGT9 A0A3B0IYZ6 A0A1W4VC90 A0A182FMZ3 A0A0J9U372 A0A1W4V0D2 A0A0P4W2W1 N6T8J1 A0A0Q5W116 Q7KN85 A0A0R1DT27 X1WV43 A0A2H8TW78 K7IWV9 B4J9W4 A0A0P8Y505 B3MC04 A0A232FD21 B4KQQ6 Q6AWP8 B3NPZ2 A0A0Q9XH45 A0A0N8P0B2 A0A0G3YKZ0 A0A336M202 A0A1I8P547 Q28WR6 A0A1I8P4X7 A0A0A9WBE8 A0A1I8P4Z2 A0A0R3NQ61 A0A0R3NVZ8 A0A0A9W2Z2 A0A0Q9XLD9 T1P9Z0 A0A0Q5VM32 A0A0Q9W5R4 B4LL37 T1HR11 B4MIX5 A0A336K4N2 B0W198 T1PC34 A0A1I8MVD7 A0A1Q3FL08 A0A1Q3FLA1 A0A026W0M5
V5GTA5 D2A5W6 A0A182U8G4 A0A182HQV2 A0A2J7RDY5 W8AYI0 A0A1A9XP97 A0A182VKF6 A0A182XLF6 A0A2J7RDZ4 A0A0A1X3Z2 A0A067QJK7 A0A1B0CWI3 A0A0V0G5Z5 A0A182PCH9 A0A224XIL5 A0A182R7Q4 A0A182WA54 A0A034VWU5 A0A1L8E1A7 A0A182Q238 U5EZ83 A0A2M3ZZ32 A0A1A9W2F7 A0A0J9RDY6 A0A1L8E112 A0A2M3ZZ25 A0A0K8U6N3 B4P6T1 A0A0L0CF01 W5J1A9 A0A2M3YY31 A0A182NPW4 A0A0B4LFH8 A0A1W4UZZ4 A0A1L8E0T3 A0A182JI57 A0A182YIL9 B4QHD4 A0A0M4EUF0 A0A182K9C8 B4HSH9 A0A182MUM6 A0A0R1DSP7 A0A069DXN3 A0A2M4BBA3 E2QCF1 A0A1A9ZGT9 A0A3B0IYZ6 A0A1W4VC90 A0A182FMZ3 A0A0J9U372 A0A1W4V0D2 A0A0P4W2W1 N6T8J1 A0A0Q5W116 Q7KN85 A0A0R1DT27 X1WV43 A0A2H8TW78 K7IWV9 B4J9W4 A0A0P8Y505 B3MC04 A0A232FD21 B4KQQ6 Q6AWP8 B3NPZ2 A0A0Q9XH45 A0A0N8P0B2 A0A0G3YKZ0 A0A336M202 A0A1I8P547 Q28WR6 A0A1I8P4X7 A0A0A9WBE8 A0A1I8P4Z2 A0A0R3NQ61 A0A0R3NVZ8 A0A0A9W2Z2 A0A0Q9XLD9 T1P9Z0 A0A0Q5VM32 A0A0Q9W5R4 B4LL37 T1HR11 B4MIX5 A0A336K4N2 B0W198 T1PC34 A0A1I8MVD7 A0A1Q3FL08 A0A1Q3FLA1 A0A026W0M5
EC Number
2.3.3.8
Pubmed
23622113
26354079
28004739
18362917
19820115
24495485
+ More
25830018 24845553 25348373 22936249 17994087 17550304 26108605 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25244985 26334808 27129103 23537049 26109357 26109356 20075255 28648823 18057021 25702953 15632085 25401762 26823975 23185243 25315136 24508170 30249741
25830018 24845553 25348373 22936249 17994087 17550304 26108605 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25244985 26334808 27129103 23537049 26109357 26109356 20075255 28648823 18057021 25702953 15632085 25401762 26823975 23185243 25315136 24508170 30249741
EMBL
GAIX01004302
JAA88258.1
KQ459348
KPJ01339.1
KQ460500
KPJ14207.1
+ More
GEZM01055134 JAV73115.1 GALX01005028 JAB63438.1 KQ971345 EFA05014.1 APCN01001603 NEVH01005280 PNF39044.1 GAMC01016623 GAMC01016622 JAB89933.1 PNF39050.1 GBXI01008228 GBXI01002581 JAD06064.1 JAD11711.1 KK853408 KDR07798.1 AJWK01032481 AJWK01032482 GECL01002654 JAP03470.1 GFTR01008545 JAW07881.1 GAKP01011181 GAKP01011180 JAC47771.1 GFDF01001732 JAV12352.1 AXCN02001998 AXCN02001999 GANO01001258 JAB58613.1 GGFK01000438 MBW33759.1 CM002911 KMY94136.1 GFDF01001730 JAV12354.1 GGFK01000440 MBW33761.1 GDHF01033454 GDHF01030000 JAI18860.1 JAI22314.1 CM000158 EDW92008.1 JRES01000487 KNC30821.1 ADMH02002186 ETN57927.1 GGFM01000410 MBW21161.1 AE013599 AHN56267.1 GFDF01001731 JAV12353.1 CM000362 EDX07279.1 KMY94133.1 CP012524 ALC41312.1 CH480816 EDW48057.1 AXCM01000963 KRK00246.1 GBGD01000239 JAC88650.1 GGFJ01001178 MBW50319.1 AAM70940.2 OUUW01000001 SPP73275.1 KMY94135.1 GDKW01000756 JAI55839.1 APGK01047658 APGK01047659 APGK01047660 APGK01047661 KB741084 ENN74028.1 CH954179 KQS62488.1 AF132166 AAD34754.2 AAF58082.2 KRK00245.1 ABLF02035281 ABLF02035282 ABLF02035291 GFXV01005613 MBW17418.1 AAZX01008339 CH916367 EDW02551.1 CH902619 KPU76635.1 EDV37191.1 NNAY01000459 OXU28247.1 CH933808 EDW09255.1 BT015200 AAT94429.1 EDV55839.1 KRG04548.1 KPU76636.1 KM507097 AKM28421.1 UFQS01000101 UFQT01000101 SSW99618.1 SSX19998.1 CM000071 EAL26601.2 GBHO01041446 GBHO01041444 GBRD01005939 GBRD01005938 GDHC01002781 JAG02158.1 JAG02160.1 JAG59882.1 JAQ15848.1 KRT03207.1 KRT03205.1 GBHO01041445 JAG02159.1 KRG04547.1 KA645494 AFP60123.1 KQS62487.1 CH940648 KRF80280.1 EDW61844.1 ACPB03019200 CH963719 EDW72064.1 SSW99619.1 SSX19999.1 DS231821 EDS44968.1 KA645493 AFP60122.1 GFDL01006807 JAV28238.1 GFDL01006789 JAV28256.1 KK107597 QOIP01000013 EZA48609.1 RLU15385.1
GEZM01055134 JAV73115.1 GALX01005028 JAB63438.1 KQ971345 EFA05014.1 APCN01001603 NEVH01005280 PNF39044.1 GAMC01016623 GAMC01016622 JAB89933.1 PNF39050.1 GBXI01008228 GBXI01002581 JAD06064.1 JAD11711.1 KK853408 KDR07798.1 AJWK01032481 AJWK01032482 GECL01002654 JAP03470.1 GFTR01008545 JAW07881.1 GAKP01011181 GAKP01011180 JAC47771.1 GFDF01001732 JAV12352.1 AXCN02001998 AXCN02001999 GANO01001258 JAB58613.1 GGFK01000438 MBW33759.1 CM002911 KMY94136.1 GFDF01001730 JAV12354.1 GGFK01000440 MBW33761.1 GDHF01033454 GDHF01030000 JAI18860.1 JAI22314.1 CM000158 EDW92008.1 JRES01000487 KNC30821.1 ADMH02002186 ETN57927.1 GGFM01000410 MBW21161.1 AE013599 AHN56267.1 GFDF01001731 JAV12353.1 CM000362 EDX07279.1 KMY94133.1 CP012524 ALC41312.1 CH480816 EDW48057.1 AXCM01000963 KRK00246.1 GBGD01000239 JAC88650.1 GGFJ01001178 MBW50319.1 AAM70940.2 OUUW01000001 SPP73275.1 KMY94135.1 GDKW01000756 JAI55839.1 APGK01047658 APGK01047659 APGK01047660 APGK01047661 KB741084 ENN74028.1 CH954179 KQS62488.1 AF132166 AAD34754.2 AAF58082.2 KRK00245.1 ABLF02035281 ABLF02035282 ABLF02035291 GFXV01005613 MBW17418.1 AAZX01008339 CH916367 EDW02551.1 CH902619 KPU76635.1 EDV37191.1 NNAY01000459 OXU28247.1 CH933808 EDW09255.1 BT015200 AAT94429.1 EDV55839.1 KRG04548.1 KPU76636.1 KM507097 AKM28421.1 UFQS01000101 UFQT01000101 SSW99618.1 SSX19998.1 CM000071 EAL26601.2 GBHO01041446 GBHO01041444 GBRD01005939 GBRD01005938 GDHC01002781 JAG02158.1 JAG02160.1 JAG59882.1 JAQ15848.1 KRT03207.1 KRT03205.1 GBHO01041445 JAG02159.1 KRG04547.1 KA645494 AFP60123.1 KQS62487.1 CH940648 KRF80280.1 EDW61844.1 ACPB03019200 CH963719 EDW72064.1 SSW99619.1 SSX19999.1 DS231821 EDS44968.1 KA645493 AFP60122.1 GFDL01006807 JAV28238.1 GFDL01006789 JAV28256.1 KK107597 QOIP01000013 EZA48609.1 RLU15385.1
Proteomes
UP000053268
UP000053240
UP000192223
UP000007266
UP000075902
UP000075840
+ More
UP000235965 UP000092443 UP000075903 UP000076407 UP000027135 UP000092461 UP000075885 UP000075900 UP000075920 UP000075886 UP000091820 UP000002282 UP000037069 UP000000673 UP000075884 UP000000803 UP000192221 UP000075880 UP000076408 UP000000304 UP000092553 UP000075881 UP000001292 UP000075883 UP000092445 UP000268350 UP000069272 UP000019118 UP000008711 UP000007819 UP000002358 UP000001070 UP000007801 UP000215335 UP000009192 UP000095300 UP000001819 UP000008792 UP000015103 UP000007798 UP000002320 UP000095301 UP000053097 UP000279307
UP000235965 UP000092443 UP000075903 UP000076407 UP000027135 UP000092461 UP000075885 UP000075900 UP000075920 UP000075886 UP000091820 UP000002282 UP000037069 UP000000673 UP000075884 UP000000803 UP000192221 UP000075880 UP000076408 UP000000304 UP000092553 UP000075881 UP000001292 UP000075883 UP000092445 UP000268350 UP000069272 UP000019118 UP000008711 UP000007819 UP000002358 UP000001070 UP000007801 UP000215335 UP000009192 UP000095300 UP000001819 UP000008792 UP000015103 UP000007798 UP000002320 UP000095301 UP000053097 UP000279307
Pfam
Interpro
IPR033847
Citrt_syn/SCS-alpha_CS
+ More
IPR003781 CoA-bd
IPR013815 ATP_grasp_subdomain_1
IPR002020 Citrate_synthase
IPR016143 Citrate_synth-like_sm_a-sub
IPR014608 ATP-citrate_synthase
IPR017866 Succ-CoA_synthase_bsu_CS
IPR032263 Citrate-bd
IPR005811 CoA_ligase
IPR016102 Succinyl-CoA_synth-like
IPR036291 NAD(P)-bd_dom_sf
IPR036969 Citrate_synthase_sf
IPR016142 Citrate_synth-like_lrg_a-sub
IPR013650 ATP-grasp_succ-CoA_synth-type
IPR017440 Cit_synth/succinyl-CoA_lig_AS
IPR003781 CoA-bd
IPR013815 ATP_grasp_subdomain_1
IPR002020 Citrate_synthase
IPR016143 Citrate_synth-like_sm_a-sub
IPR014608 ATP-citrate_synthase
IPR017866 Succ-CoA_synthase_bsu_CS
IPR032263 Citrate-bd
IPR005811 CoA_ligase
IPR016102 Succinyl-CoA_synth-like
IPR036291 NAD(P)-bd_dom_sf
IPR036969 Citrate_synthase_sf
IPR016142 Citrate_synth-like_lrg_a-sub
IPR013650 ATP-grasp_succ-CoA_synth-type
IPR017440 Cit_synth/succinyl-CoA_lig_AS
ProteinModelPortal
S4PEX5
A0A0N0P9L9
A0A194R999
A0A1Y1LMG7
A0A1W4WDM4
A0A1W4WEX6
+ More
V5GTA5 D2A5W6 A0A182U8G4 A0A182HQV2 A0A2J7RDY5 W8AYI0 A0A1A9XP97 A0A182VKF6 A0A182XLF6 A0A2J7RDZ4 A0A0A1X3Z2 A0A067QJK7 A0A1B0CWI3 A0A0V0G5Z5 A0A182PCH9 A0A224XIL5 A0A182R7Q4 A0A182WA54 A0A034VWU5 A0A1L8E1A7 A0A182Q238 U5EZ83 A0A2M3ZZ32 A0A1A9W2F7 A0A0J9RDY6 A0A1L8E112 A0A2M3ZZ25 A0A0K8U6N3 B4P6T1 A0A0L0CF01 W5J1A9 A0A2M3YY31 A0A182NPW4 A0A0B4LFH8 A0A1W4UZZ4 A0A1L8E0T3 A0A182JI57 A0A182YIL9 B4QHD4 A0A0M4EUF0 A0A182K9C8 B4HSH9 A0A182MUM6 A0A0R1DSP7 A0A069DXN3 A0A2M4BBA3 E2QCF1 A0A1A9ZGT9 A0A3B0IYZ6 A0A1W4VC90 A0A182FMZ3 A0A0J9U372 A0A1W4V0D2 A0A0P4W2W1 N6T8J1 A0A0Q5W116 Q7KN85 A0A0R1DT27 X1WV43 A0A2H8TW78 K7IWV9 B4J9W4 A0A0P8Y505 B3MC04 A0A232FD21 B4KQQ6 Q6AWP8 B3NPZ2 A0A0Q9XH45 A0A0N8P0B2 A0A0G3YKZ0 A0A336M202 A0A1I8P547 Q28WR6 A0A1I8P4X7 A0A0A9WBE8 A0A1I8P4Z2 A0A0R3NQ61 A0A0R3NVZ8 A0A0A9W2Z2 A0A0Q9XLD9 T1P9Z0 A0A0Q5VM32 A0A0Q9W5R4 B4LL37 T1HR11 B4MIX5 A0A336K4N2 B0W198 T1PC34 A0A1I8MVD7 A0A1Q3FL08 A0A1Q3FLA1 A0A026W0M5
V5GTA5 D2A5W6 A0A182U8G4 A0A182HQV2 A0A2J7RDY5 W8AYI0 A0A1A9XP97 A0A182VKF6 A0A182XLF6 A0A2J7RDZ4 A0A0A1X3Z2 A0A067QJK7 A0A1B0CWI3 A0A0V0G5Z5 A0A182PCH9 A0A224XIL5 A0A182R7Q4 A0A182WA54 A0A034VWU5 A0A1L8E1A7 A0A182Q238 U5EZ83 A0A2M3ZZ32 A0A1A9W2F7 A0A0J9RDY6 A0A1L8E112 A0A2M3ZZ25 A0A0K8U6N3 B4P6T1 A0A0L0CF01 W5J1A9 A0A2M3YY31 A0A182NPW4 A0A0B4LFH8 A0A1W4UZZ4 A0A1L8E0T3 A0A182JI57 A0A182YIL9 B4QHD4 A0A0M4EUF0 A0A182K9C8 B4HSH9 A0A182MUM6 A0A0R1DSP7 A0A069DXN3 A0A2M4BBA3 E2QCF1 A0A1A9ZGT9 A0A3B0IYZ6 A0A1W4VC90 A0A182FMZ3 A0A0J9U372 A0A1W4V0D2 A0A0P4W2W1 N6T8J1 A0A0Q5W116 Q7KN85 A0A0R1DT27 X1WV43 A0A2H8TW78 K7IWV9 B4J9W4 A0A0P8Y505 B3MC04 A0A232FD21 B4KQQ6 Q6AWP8 B3NPZ2 A0A0Q9XH45 A0A0N8P0B2 A0A0G3YKZ0 A0A336M202 A0A1I8P547 Q28WR6 A0A1I8P4X7 A0A0A9WBE8 A0A1I8P4Z2 A0A0R3NQ61 A0A0R3NVZ8 A0A0A9W2Z2 A0A0Q9XLD9 T1P9Z0 A0A0Q5VM32 A0A0Q9W5R4 B4LL37 T1HR11 B4MIX5 A0A336K4N2 B0W198 T1PC34 A0A1I8MVD7 A0A1Q3FL08 A0A1Q3FLA1 A0A026W0M5
PDB
6O0H
E-value=0,
Score=3531
Ontologies
PATHWAY
GO
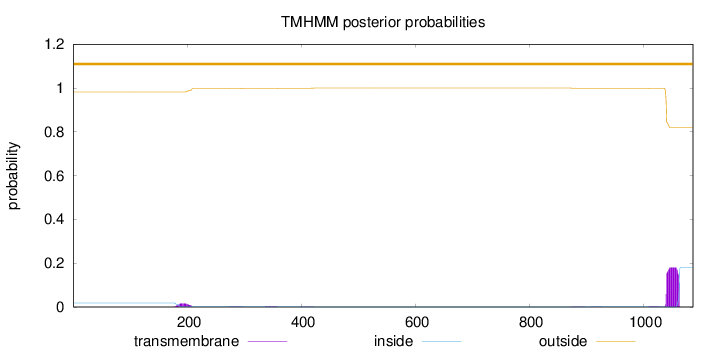
Topology
Subcellular location
Cytoplasm
Length:
1087
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.45418
Exp number, first 60 AAs:
0.00199
Total prob of N-in:
0.01841
outside
1 - 1087
Population Genetic Test Statistics
Pi
318.112222
Theta
181.291794
Tajima's D
2.808848
CLR
0.210824
CSRT
0.968201589920504
Interpretation
Uncertain
