Gene
KWMTBOMO14532
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.306
Sequence
CDS
ATGGTCAGGAACGACAGGCTCTCTGCTCGTGGTGGTGGTACCGTCATTTACTATAGAAGAGCCCTGCATTGCGTCCCGCTCGATCCTCCCGCGCTCGCTAATATCGAAGCATCAGTGTGCCGAATCTCACTGACGGGGCACGCGCCGATCGTTATCGCGTCCGTTTATCTTCCACCGGATAAGATCGTTCTAAGCAGTGATATCGAGGCACTGCTCGGCATGGGGAGCTCTGTCATTCTGGCGGGCGACCTAAATTGTAAACACATCAGGTGGAACTCACACACCACAACCCCGAATGGCAGGCGGCTTGACGCGTTAGTCGATGATCTCGCCTTCGATATCGTCGCTCCGCTAACACCGACTCACTACCCGCTAAATATCGCGCATCGCCCGGATGTACTCGACATAGCGTTATTAAAAAACGTAACTCTGAGCTTACACTCGATCGAAGTAGTTTCAGAGTTAGATTCAGACCACCGTCCCGTCGTTATGAAGCTCGGTCGCGCTCCCGACTCCGTTCCCGTCACGAGGACTGTGGTGGATTGGCACACGCTGGGCATCAGCCTGGCTGAATCTGATCCACCATCGCTCCCGTTTAACCCGGACTCTATCCCGTCTCCTCAGGATACCGCTGAAGCCATAGACATCTTAACGTCACACATCACCTCAACATTAGATAGATCATCGAAACAAGTTGTAGCGGAGGATTTCCTTCACCGCTTCAAATTGTCCGATGATATTAGGGAACTCCTTAGAGCTAAGAACGCCTCGATACGCGCCTACGACAGGTATCCTACCGCGGAAAATCGTATTCGAATGCGTGCCCTACAACGCGACGTAAAGTCTCGCATCGCCGAAGTCCGAGATGCCAGATGGTCTGATTTCTTAGAAGGACTCGCGCCCTCCCAAAGGTCTTACTACCGCTTAGCTCGTACTCTCAAATCGGATACGGTAGTAACTATGCCCCCCCTCGTAGGCCCCTCAGGCCGACTCGCGGCGTTTGATGATGACGAAAAAGCAGAGTTGCTGGCCGATACATTGCAAACCCAGTGCACGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGTCTCCTTGCCACCCTCGGATGCGTTACCACCCGTCACCCCGATGGAAGTTAAGGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACGGTATATCCAACCGCGTTATTAAACTTCTGCCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGCATACATAAACCAGGTAAACCAAAAAATCATCCGACGAGCTACCGCCCGATTAGCCTCCTCATGTCTCTAG
Protein
MVRNDRLSARGGGTVIYYRRALHCVPLDPPALANIEASVCRISLTGHAPIVIASVYLPPDKIVLSSDIEALLGMGSSVILAGDLNCKHIRWNSHTTTPNGRRLDALVDDLAFDIVAPLTPTHYPLNIAHRPDVLDIALLKNVTLSLHSIEVVSELDSDHRPVVMKLGRAPDSVPVTRTVVDWHTLGISLAESDPPSLPFNPDSIPSPQDTAEAIDILTSHITSTLDRSSKQVVAEDFLHRFKLSDDIRELLRAKNASIRAYDRYPTAENRIRMRALQRDVKSRIAEVRDARWSDFLEGLAPSQRSYYRLARTLKSDTVVTMPPLVGPSGRLAAFDDDEKAELLADTLQTQCTPSTQSVDPVHVELVDSEVERRVSLPPSDALPPVTPMEVKDLIKDLRPRKAPGSDGISNRVIKLLPVQLIVMLASIFNAAMANCIFPRCGKKRTLSAYINQVNQKIIRRATARLASSCL
Summary
Uniprot
Q93137
Q9BPQ0
Q9XXW0
A0A0N1PFV1
O96546
A0A1Y1NA23
+ More
A0A139W8G9 K7IN67 V5GNJ2 A0A1B6MIM7 A0A139WA11 X1X0M4 A0A2S2QEE0 A0A023EYS8 A0A087THU2 J9LC21 A0A2S2PEZ9 A0A087UCW3 A0A087U3K8 A0A2S2NJB5 A0A087T3W0 X1WQI9 A0A2S2P6H1 V5GNF3 A0A1Y1K0C8 A0A224XB44 X1WNI2 X1WK15 X1WLB2 A0A2S2N6Q7 T1ID59 A0A224X6Q4 A0A2S2PPK8 X1WKS2 U4UGQ8 X1WVL6 A0A2J7PP17 J9LFC9 A0A087UB64 J9KPF1 X1WZH0 J9KKL6 A0A224XI99 A0A2M4AQV0 A0A2M4AQK3 A0A224X6T4 A0A2H8TF37 A0A224X6S1 A0A2S2PJ52 A0A1B6C2V8 J9MAR0 A0A1Y1KFT3 A0A069DZL8 A0A2S2PA10 A0A2S2P6S3 A0A1Y1KFR3 A0A224X6U5 A0A224X6W2 J9JS95 A0A224XJI0 J9LVT4 A0A2M4CV42 A0A2J7R7D0 A0A2A4IUM2 A0A2M4CV52 A0A2S2NCK2 A0A2S2NH02 J9JZG6 J9M522 A0A0V0G5B6 A0A2S2R7N0 A0A2S2Q383 A0A2S2R2E8 A0A087UGF5 A0A2S2QV22 J9MAC6 A0A2J7PQB5 A0A2S2QDN9 A0A0J7K503 A0A023F1L2 A0A2S2Q2R8 A0A2S2PJG2 A0A3S2NJ07 A0A232ENU1 A0A224X746 J9KLB1 A0A023F0I3 A0A224X724 A0A023EZE6 A0A2S2P7B2 A0A1Y1LKN6 J9LBF1 A0A0J7K0R9 A0A224XF24 A0A224X7Z3 A0A0P4VK54 A0A2J7QR37 A0A2J7PBY3 A0A224XIB0
A0A139W8G9 K7IN67 V5GNJ2 A0A1B6MIM7 A0A139WA11 X1X0M4 A0A2S2QEE0 A0A023EYS8 A0A087THU2 J9LC21 A0A2S2PEZ9 A0A087UCW3 A0A087U3K8 A0A2S2NJB5 A0A087T3W0 X1WQI9 A0A2S2P6H1 V5GNF3 A0A1Y1K0C8 A0A224XB44 X1WNI2 X1WK15 X1WLB2 A0A2S2N6Q7 T1ID59 A0A224X6Q4 A0A2S2PPK8 X1WKS2 U4UGQ8 X1WVL6 A0A2J7PP17 J9LFC9 A0A087UB64 J9KPF1 X1WZH0 J9KKL6 A0A224XI99 A0A2M4AQV0 A0A2M4AQK3 A0A224X6T4 A0A2H8TF37 A0A224X6S1 A0A2S2PJ52 A0A1B6C2V8 J9MAR0 A0A1Y1KFT3 A0A069DZL8 A0A2S2PA10 A0A2S2P6S3 A0A1Y1KFR3 A0A224X6U5 A0A224X6W2 J9JS95 A0A224XJI0 J9LVT4 A0A2M4CV42 A0A2J7R7D0 A0A2A4IUM2 A0A2M4CV52 A0A2S2NCK2 A0A2S2NH02 J9JZG6 J9M522 A0A0V0G5B6 A0A2S2R7N0 A0A2S2Q383 A0A2S2R2E8 A0A087UGF5 A0A2S2QV22 J9MAC6 A0A2J7PQB5 A0A2S2QDN9 A0A0J7K503 A0A023F1L2 A0A2S2Q2R8 A0A2S2PJG2 A0A3S2NJ07 A0A232ENU1 A0A224X746 J9KLB1 A0A023F0I3 A0A224X724 A0A023EZE6 A0A2S2P7B2 A0A1Y1LKN6 J9LBF1 A0A0J7K0R9 A0A224XF24 A0A224X7Z3 A0A0P4VK54 A0A2J7QR37 A0A2J7PBY3 A0A224XIB0
Pubmed
EMBL
U07847
AAA17752.1
AB055391
BAB21761.1
AB018558
BAA76304.1
+ More
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 KQ973342 KXZ75569.1 GALX01005299 JAB63167.1 GEBQ01004157 JAT35820.1 KQ971729 KYB24751.1 ABLF02065839 GGMS01006359 MBY75562.1 GBBI01004395 JAC14317.1 KK115285 KFM64681.1 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 GGMR01015412 MBY28031.1 KK119261 KFM75202.1 KK118005 KFM71947.1 GGMR01004661 MBY17280.1 KK113281 KFM59799.1 ABLF02022418 GGMR01012189 MBY24808.1 GALX01005359 JAB63107.1 GEZM01099921 GEZM01099920 GEZM01099918 GEZM01099906 JAV53125.1 GFTR01008262 JAW08164.1 ABLF02008153 ABLF02008462 ABLF02008464 ABLF02012584 ABLF02013765 ABLF02024884 ABLF02042518 ABLF02057746 GGMR01000226 MBY12845.1 ACPB03027378 GFTR01008279 JAW08147.1 GGMR01018716 MBY31335.1 ABLF02005245 ABLF02013477 ABLF02047385 ABLF02066435 KB632143 ERL89075.1 ABLF02042385 NEVH01023280 PNF18085.1 ABLF02023911 KK119069 KFM74603.1 ABLF02041767 ABLF02034467 ABLF02015631 ABLF02021986 ABLF02021987 ABLF02021991 GFTR01008256 JAW08170.1 GGFK01009846 MBW43167.1 GGFK01009732 MBW43053.1 GFTR01008261 JAW08165.1 GFXV01000908 MBW12713.1 GFTR01008271 JAW08155.1 GGMR01016297 MBY28916.1 GEDC01029594 JAS07704.1 ABLF02037715 GEZM01084970 JAV60359.1 GBGD01000415 JAC88474.1 GGMR01013409 MBY26028.1 GGMR01012443 MBY25062.1 GEZM01084969 JAV60363.1 GFTR01008251 JAW08175.1 GFTR01008260 JAW08166.1 ABLF02028535 ABLF02028545 GFTR01008257 JAW08169.1 ABLF02003241 ABLF02003242 ABLF02025904 GGFL01005012 MBW69190.1 NEVH01006736 PNF36716.1 NWSH01007753 PCG62823.1 GGFL01004957 MBW69135.1 GGMR01002322 MBY14941.1 GGMR01003864 MBY16483.1 ABLF02037436 ABLF02024246 GECL01003538 JAP02586.1 GGMS01016818 MBY86021.1 GGMS01002992 MBY72195.1 GGMS01014910 MBY84113.1 KK119692 KFM76444.1 GGMS01012398 MBY81601.1 ABLF02007779 ABLF02007782 ABLF02050375 NEVH01022639 PNF18527.1 GGMS01006655 MBY75858.1 LBMM01014024 KMQ85369.1 GBBI01003811 JAC14901.1 GGMS01002786 MBY71989.1 GGMR01016905 MBY29524.1 RSAL01000708 RVE41206.1 NNAY01003095 OXU19992.1 GFTR01008151 JAW08275.1 ABLF02040174 ABLF02040183 ABLF02055266 GBBI01003747 JAC14965.1 GFTR01008169 JAW08257.1 GBBI01004571 JAC14141.1 GGMR01012676 MBY25295.1 GEZM01053022 JAV74219.1 ABLF02012739 ABLF02012741 LBMM01018021 KMQ83899.1 GFTR01008038 JAW08388.1 GFTR01007929 JAW08497.1 GDKW01003730 JAI52865.1 NEVH01011985 PNF31054.1 NEVH01027074 PNF13841.1 GFTR01008246 JAW08180.1
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 KQ973342 KXZ75569.1 GALX01005299 JAB63167.1 GEBQ01004157 JAT35820.1 KQ971729 KYB24751.1 ABLF02065839 GGMS01006359 MBY75562.1 GBBI01004395 JAC14317.1 KK115285 KFM64681.1 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 GGMR01015412 MBY28031.1 KK119261 KFM75202.1 KK118005 KFM71947.1 GGMR01004661 MBY17280.1 KK113281 KFM59799.1 ABLF02022418 GGMR01012189 MBY24808.1 GALX01005359 JAB63107.1 GEZM01099921 GEZM01099920 GEZM01099918 GEZM01099906 JAV53125.1 GFTR01008262 JAW08164.1 ABLF02008153 ABLF02008462 ABLF02008464 ABLF02012584 ABLF02013765 ABLF02024884 ABLF02042518 ABLF02057746 GGMR01000226 MBY12845.1 ACPB03027378 GFTR01008279 JAW08147.1 GGMR01018716 MBY31335.1 ABLF02005245 ABLF02013477 ABLF02047385 ABLF02066435 KB632143 ERL89075.1 ABLF02042385 NEVH01023280 PNF18085.1 ABLF02023911 KK119069 KFM74603.1 ABLF02041767 ABLF02034467 ABLF02015631 ABLF02021986 ABLF02021987 ABLF02021991 GFTR01008256 JAW08170.1 GGFK01009846 MBW43167.1 GGFK01009732 MBW43053.1 GFTR01008261 JAW08165.1 GFXV01000908 MBW12713.1 GFTR01008271 JAW08155.1 GGMR01016297 MBY28916.1 GEDC01029594 JAS07704.1 ABLF02037715 GEZM01084970 JAV60359.1 GBGD01000415 JAC88474.1 GGMR01013409 MBY26028.1 GGMR01012443 MBY25062.1 GEZM01084969 JAV60363.1 GFTR01008251 JAW08175.1 GFTR01008260 JAW08166.1 ABLF02028535 ABLF02028545 GFTR01008257 JAW08169.1 ABLF02003241 ABLF02003242 ABLF02025904 GGFL01005012 MBW69190.1 NEVH01006736 PNF36716.1 NWSH01007753 PCG62823.1 GGFL01004957 MBW69135.1 GGMR01002322 MBY14941.1 GGMR01003864 MBY16483.1 ABLF02037436 ABLF02024246 GECL01003538 JAP02586.1 GGMS01016818 MBY86021.1 GGMS01002992 MBY72195.1 GGMS01014910 MBY84113.1 KK119692 KFM76444.1 GGMS01012398 MBY81601.1 ABLF02007779 ABLF02007782 ABLF02050375 NEVH01022639 PNF18527.1 GGMS01006655 MBY75858.1 LBMM01014024 KMQ85369.1 GBBI01003811 JAC14901.1 GGMS01002786 MBY71989.1 GGMR01016905 MBY29524.1 RSAL01000708 RVE41206.1 NNAY01003095 OXU19992.1 GFTR01008151 JAW08275.1 ABLF02040174 ABLF02040183 ABLF02055266 GBBI01003747 JAC14965.1 GFTR01008169 JAW08257.1 GBBI01004571 JAC14141.1 GGMR01012676 MBY25295.1 GEZM01053022 JAV74219.1 ABLF02012739 ABLF02012741 LBMM01018021 KMQ83899.1 GFTR01008038 JAW08388.1 GFTR01007929 JAW08497.1 GDKW01003730 JAI52865.1 NEVH01011985 PNF31054.1 NEVH01027074 PNF13841.1 GFTR01008246 JAW08180.1
Proteomes
Pfam
Interpro
IPR036691
Endo/exonu/phosph_ase_sf
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR006579 Pre_C2HC_dom
IPR013103 RVT_2
IPR025398 DUF4371
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR037220 BED_dom
IPR007021 DUF659
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR006579 Pre_C2HC_dom
IPR013103 RVT_2
IPR025398 DUF4371
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR037220 BED_dom
IPR007021 DUF659
Gene 3D
ProteinModelPortal
Q93137
Q9BPQ0
Q9XXW0
A0A0N1PFV1
O96546
A0A1Y1NA23
+ More
A0A139W8G9 K7IN67 V5GNJ2 A0A1B6MIM7 A0A139WA11 X1X0M4 A0A2S2QEE0 A0A023EYS8 A0A087THU2 J9LC21 A0A2S2PEZ9 A0A087UCW3 A0A087U3K8 A0A2S2NJB5 A0A087T3W0 X1WQI9 A0A2S2P6H1 V5GNF3 A0A1Y1K0C8 A0A224XB44 X1WNI2 X1WK15 X1WLB2 A0A2S2N6Q7 T1ID59 A0A224X6Q4 A0A2S2PPK8 X1WKS2 U4UGQ8 X1WVL6 A0A2J7PP17 J9LFC9 A0A087UB64 J9KPF1 X1WZH0 J9KKL6 A0A224XI99 A0A2M4AQV0 A0A2M4AQK3 A0A224X6T4 A0A2H8TF37 A0A224X6S1 A0A2S2PJ52 A0A1B6C2V8 J9MAR0 A0A1Y1KFT3 A0A069DZL8 A0A2S2PA10 A0A2S2P6S3 A0A1Y1KFR3 A0A224X6U5 A0A224X6W2 J9JS95 A0A224XJI0 J9LVT4 A0A2M4CV42 A0A2J7R7D0 A0A2A4IUM2 A0A2M4CV52 A0A2S2NCK2 A0A2S2NH02 J9JZG6 J9M522 A0A0V0G5B6 A0A2S2R7N0 A0A2S2Q383 A0A2S2R2E8 A0A087UGF5 A0A2S2QV22 J9MAC6 A0A2J7PQB5 A0A2S2QDN9 A0A0J7K503 A0A023F1L2 A0A2S2Q2R8 A0A2S2PJG2 A0A3S2NJ07 A0A232ENU1 A0A224X746 J9KLB1 A0A023F0I3 A0A224X724 A0A023EZE6 A0A2S2P7B2 A0A1Y1LKN6 J9LBF1 A0A0J7K0R9 A0A224XF24 A0A224X7Z3 A0A0P4VK54 A0A2J7QR37 A0A2J7PBY3 A0A224XIB0
A0A139W8G9 K7IN67 V5GNJ2 A0A1B6MIM7 A0A139WA11 X1X0M4 A0A2S2QEE0 A0A023EYS8 A0A087THU2 J9LC21 A0A2S2PEZ9 A0A087UCW3 A0A087U3K8 A0A2S2NJB5 A0A087T3W0 X1WQI9 A0A2S2P6H1 V5GNF3 A0A1Y1K0C8 A0A224XB44 X1WNI2 X1WK15 X1WLB2 A0A2S2N6Q7 T1ID59 A0A224X6Q4 A0A2S2PPK8 X1WKS2 U4UGQ8 X1WVL6 A0A2J7PP17 J9LFC9 A0A087UB64 J9KPF1 X1WZH0 J9KKL6 A0A224XI99 A0A2M4AQV0 A0A2M4AQK3 A0A224X6T4 A0A2H8TF37 A0A224X6S1 A0A2S2PJ52 A0A1B6C2V8 J9MAR0 A0A1Y1KFT3 A0A069DZL8 A0A2S2PA10 A0A2S2P6S3 A0A1Y1KFR3 A0A224X6U5 A0A224X6W2 J9JS95 A0A224XJI0 J9LVT4 A0A2M4CV42 A0A2J7R7D0 A0A2A4IUM2 A0A2M4CV52 A0A2S2NCK2 A0A2S2NH02 J9JZG6 J9M522 A0A0V0G5B6 A0A2S2R7N0 A0A2S2Q383 A0A2S2R2E8 A0A087UGF5 A0A2S2QV22 J9MAC6 A0A2J7PQB5 A0A2S2QDN9 A0A0J7K503 A0A023F1L2 A0A2S2Q2R8 A0A2S2PJG2 A0A3S2NJ07 A0A232ENU1 A0A224X746 J9KLB1 A0A023F0I3 A0A224X724 A0A023EZE6 A0A2S2P7B2 A0A1Y1LKN6 J9LBF1 A0A0J7K0R9 A0A224XF24 A0A224X7Z3 A0A0P4VK54 A0A2J7QR37 A0A2J7PBY3 A0A224XIB0
Ontologies
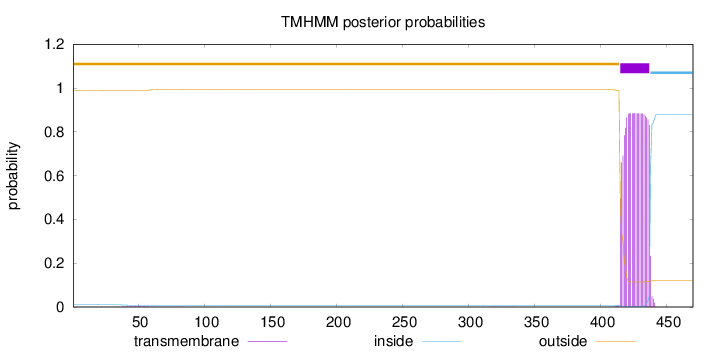
Topology
Length:
470
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.66509
Exp number, first 60 AAs:
0.05517
Total prob of N-in:
0.01069
outside
1 - 414
TMhelix
415 - 437
inside
438 - 470
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0.000539
CSRT
0
Interpretation
Uncertain
