Gene
KWMTBOMO14530 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014398
Annotation
PREDICTED:_eukaryotic_peptide_chain_release_factor_subunit_1_[Amyelois_transitella]
Full name
Eukaryotic peptide chain release factor subunit 1
Location in the cell
Cytoplasmic Reliability : 3.269
Sequence
CDS
ATGTCTGAAGAATCGTCGGCCGATCGAAATGTCGAAATATGGAAGATAAAGAAACTTATAAAGAGTTTGGAAATGGCCAGAGGAAATGGAACATCGATGATATCCCTGATCATTCCACCTAAGGATCAAATCTCGAGAGTATCAAAGATGTTGGCTGATGAATTTGGTACAGCTTCTAATATCAAGTCACGTGTGAATCGTCTCTCAGTGCTTGGCGCAATTACTTCTGTACAGCACCGACTCAAGCTGTATACTAAAGTGCCACCAAACGGCCTAGTTATATACTGTGGTACTATCGTGACAGAAGAAGGTAAAGAGAAGAAAGTGAACATTGATTTTGAGCCGTTCAAGCCTATCAACACATCTCTTTATCTCTGCGACAACAAGTTCCACACCGAGGCTCTAACAGCACTGCTGGCTGATGATAACAAGTTTGGTTTCATTGTTATGGATGGTAACGGTGCATTATTCGGTACCCTTCAAGGAAACACTAGAGAGGTACTCCACAAGTTTACTGTCGATCTCCCTAAGAAGCACGGTCGCGGTGGTCAGTCAGCGCTGCGTTTCGCTCGTCTCCGTATGGAGAAACGTCACAATTACGTCCGTAAGGTTGCCGAGGTAGCCACCCAGCTGTTCATCAGTGCTGACCGGCCGAACGTTGCTGGCCTCATCCTTGCGGGTTCTGCTGATTTTAAGACTGAACTGTCCCAGTCCGATATGTTTGATCCGCGTCTCCAAGCCAAGATCATAAAACTGGTGGACGTATCATATGGAGGCGAAAACGGTTTCAATCAGGCTATAGAGCTGGCTGCGGAATCCTTGCAGAATGTCAAGTTTATACAGGAGAAGAAACTAATTGGACGCTACTTTGATGAGATCTCACAGGACACAGGAAAGTATTGCTTCGGTGTCGATGATACACTGCGCGCTCTCGAACTCGGATCCGTCGAGACGTTGATCTGCTGGGAGAACCTCGACATACAAAGATACGTATTGAAGTCGCATGCCACCAACCAGGAAACAATTCTCCACCTGACGCCTGAACAAGAGAAGGACAAATCACATTTCACCGACAAAGAGAGCGGAGTGGAGCTGGAGCTTGTGGAGTGCCAGCCGCTGTTGGAGTGGCTGGCCAATAACTACAAGTCGTTTGGTGCCACCTTAGAGATCATCACGGACAAGAGCCAGGAGGGAAGCCAGTTCGTTCGCGGCTTCGGAGGGATCGGCGGTCTCCTCCGTTACAAAGTGGACTTCCAATCGATGCAGCTCGACGACGAGGAAATCGACAATCTATACGACATCGACGACTATTAG
Protein
MSEESSADRNVEIWKIKKLIKSLEMARGNGTSMISLIIPPKDQISRVSKMLADEFGTASNIKSRVNRLSVLGAITSVQHRLKLYTKVPPNGLVIYCGTIVTEEGKEKKVNIDFEPFKPINTSLYLCDNKFHTEALTALLADDNKFGFIVMDGNGALFGTLQGNTREVLHKFTVDLPKKHGRGGQSALRFARLRMEKRHNYVRKVAEVATQLFISADRPNVAGLILAGSADFKTELSQSDMFDPRLQAKIIKLVDVSYGGENGFNQAIELAAESLQNVKFIQEKKLIGRYFDEISQDTGKYCFGVDDTLRALELGSVETLICWENLDIQRYVLKSHATNQETILHLTPEQEKDKSHFTDKESGVELELVECQPLLEWLANNYKSFGATLEIITDKSQEGSQFVRGFGGIGGLLRYKVDFQSMQLDDEEIDNLYDIDDY
Summary
Description
Directs the termination of nascent peptide synthesis (translation) in response to the termination codons UAA, UAG and UGA.
Subunit
Heterodimer of two subunits, one of which binds GTP.
Similarity
Belongs to the eukaryotic release factor 1 family.
Keywords
Complete proteome
Cytoplasm
Phosphoprotein
Protein biosynthesis
Reference proteome
Feature
chain Eukaryotic peptide chain release factor subunit 1
Uniprot
I4DNA6
A0A3S2M1H9
S4P221
A0A1E1WMS9
Q2F5R7
A0A194R8P0
+ More
A0A194PWL8 A0A2A4JDH0 A0A232EU20 K7IXJ8 E2A5U8 A0A026WXX0 A0A2A3EM31 A0A154PQ89 A0A195DKK3 F4WBG5 A0A195B3F9 A0A195FTR3 A0A158P126 A0A2R7VYF7 A0A0A9Y8A5 A0A224XCL0 A0A2J7RMF9 A0A0T6B7W7 R4WRS9 A0A0P4VUH7 T1HR39 A0A067QX62 A0A069DZ03 A0A0A1WNW3 A0A1Y1MM32 A0A1B6DYM6 A0A1B6HQR2 V5I9K4 J9JYP0 A0A2S2R218 A0A2S2N8H0 A0A0K8TNX1 W8BUE3 N6TX29 A0A1B6MKS2 A0A1B6FV35 A0A158P127 H9JXY0 A0A151WWM3 A0A1W4W6E5 A0A0J9RYL4 B4KY20 B3NIJ7 B3M975 B4PFA6 A0A0C9R7G1 A0A034W963 A0A1B0ARB1 B4GR19 A0A1A9ZFX9 D1Z373 D3TRE7 Q9VPH7 A0A0M5J8C5 B4MLK5 Q16P70 B4LGF2 A0A0K8V2R2 A0A3B0KLP6 B4IA85 A0A182W7Y2 A0A023ES91 A0A2W1BDZ4 D6WCG2 B4QRX2 A0A1Q3F6M8 B0WFV9 A0A084VJJ4 T1D417 A0A182JB29 B4IWX5 A0A182ML94 A0A182QFY0 A0A182NP75 Q7PLZ0 A0A182RS95 A0A182KE76 A0A182FLW9 A0A182PWU4 A0A182YDX7 A0A2M3ZFR8 A0A2J7RMG1 A0A2M4A4E1 W5JVA6 T1PDF9 A0A182GW91 A0A0R3P821 A0A1Q3FWQ6 A0A336KGH0 A0A034WAN3 A0A0K8S805 A0A1W4VUF2 A0A0Q9XCM5 M9PD27
A0A194PWL8 A0A2A4JDH0 A0A232EU20 K7IXJ8 E2A5U8 A0A026WXX0 A0A2A3EM31 A0A154PQ89 A0A195DKK3 F4WBG5 A0A195B3F9 A0A195FTR3 A0A158P126 A0A2R7VYF7 A0A0A9Y8A5 A0A224XCL0 A0A2J7RMF9 A0A0T6B7W7 R4WRS9 A0A0P4VUH7 T1HR39 A0A067QX62 A0A069DZ03 A0A0A1WNW3 A0A1Y1MM32 A0A1B6DYM6 A0A1B6HQR2 V5I9K4 J9JYP0 A0A2S2R218 A0A2S2N8H0 A0A0K8TNX1 W8BUE3 N6TX29 A0A1B6MKS2 A0A1B6FV35 A0A158P127 H9JXY0 A0A151WWM3 A0A1W4W6E5 A0A0J9RYL4 B4KY20 B3NIJ7 B3M975 B4PFA6 A0A0C9R7G1 A0A034W963 A0A1B0ARB1 B4GR19 A0A1A9ZFX9 D1Z373 D3TRE7 Q9VPH7 A0A0M5J8C5 B4MLK5 Q16P70 B4LGF2 A0A0K8V2R2 A0A3B0KLP6 B4IA85 A0A182W7Y2 A0A023ES91 A0A2W1BDZ4 D6WCG2 B4QRX2 A0A1Q3F6M8 B0WFV9 A0A084VJJ4 T1D417 A0A182JB29 B4IWX5 A0A182ML94 A0A182QFY0 A0A182NP75 Q7PLZ0 A0A182RS95 A0A182KE76 A0A182FLW9 A0A182PWU4 A0A182YDX7 A0A2M3ZFR8 A0A2J7RMG1 A0A2M4A4E1 W5JVA6 T1PDF9 A0A182GW91 A0A0R3P821 A0A1Q3FWQ6 A0A336KGH0 A0A034WAN3 A0A0K8S805 A0A1W4VUF2 A0A0Q9XCM5 M9PD27
Pubmed
22651552
23622113
26354079
28648823
20075255
20798317
+ More
24508170 30249741 21719571 21347285 25401762 26823975 23691247 27129103 24845553 26334808 25830018 28004739 26369729 24495485 23537049 19121390 22936249 17994087 18057021 17550304 25348373 20353571 10731132 12537572 12537569 18327897 17510324 24945155 28756777 18362917 19820115 24438588 24330624 12364791 14747013 17210077 25244985 20920257 23761445 25315136 26483478 15632085 12537568 12537573 12537574 16110336 17569856 17569867
24508170 30249741 21719571 21347285 25401762 26823975 23691247 27129103 24845553 26334808 25830018 28004739 26369729 24495485 23537049 19121390 22936249 17994087 18057021 17550304 25348373 20353571 10731132 12537572 12537569 18327897 17510324 24945155 28756777 18362917 19820115 24438588 24330624 12364791 14747013 17210077 25244985 20920257 23761445 25315136 26483478 15632085 12537568 12537573 12537574 16110336 17569856 17569867
EMBL
AK402804
BAM19396.1
RSAL01000072
RVE49045.1
GAIX01012215
JAA80345.1
+ More
GDQN01009915 GDQN01002893 JAT81139.1 JAT88161.1 DQ311356 ABD36300.1 KQ460500 KPJ14203.1 KQ459589 KPI97717.1 NWSH01001904 PCG69728.1 NNAY01002185 OXU21853.1 AAZX01004642 GL437061 EFN71190.1 KK107064 QOIP01000008 EZA60920.1 RLU19693.1 KZ288212 PBC32815.1 KQ435034 KZC14052.1 KQ980765 KYN13367.1 GL888063 EGI68444.1 KQ976641 KYM78805.1 KQ981272 KYN43853.1 ADTU01006100 ADTU01006101 KK854169 PTY12492.1 GBHO01015205 GBRD01007346 GDHC01012256 GDHC01003745 JAG28399.1 JAG58475.1 JAQ06373.1 JAQ14884.1 GFTR01006260 JAW10166.1 NEVH01002556 PNF42027.1 LJIG01009252 KRT83452.1 AK417407 BAN20622.1 GDKW01000912 JAI55683.1 ACPB03022348 KK853149 KDR10692.1 GBGD01001490 JAC87399.1 GBXI01013961 JAD00331.1 GEZM01027433 JAV86743.1 GEDC01006518 JAS30780.1 GECU01030690 JAS77016.1 GALX01002780 JAB65686.1 ABLF02032620 GGMS01014179 MBY83382.1 GGMR01000826 MBY13445.1 GDAI01001516 JAI16087.1 GAMC01013811 JAB92744.1 APGK01051689 APGK01051690 APGK01051691 APGK01051692 APGK01051693 KB741201 KB632375 ENN72951.1 ERL93875.1 GEBQ01003463 JAT36514.1 GECZ01022892 GECZ01015711 GECZ01002578 JAS46877.1 JAS54058.1 JAS67191.1 BABH01040929 BABH01040930 BABH01040931 KQ982691 KYQ52216.1 CM002912 KMZ00747.1 KMZ00748.1 KMZ00749.1 KMZ00752.1 KMZ00753.1 KMZ00754.1 KMZ00755.1 CH933809 EDW18722.1 KRG06336.1 CH954178 EDV52493.1 KQS44302.1 KQS44303.1 KQS44304.1 CH902618 EDV40059.1 KPU78351.1 KPU78352.1 CM000159 EDW95192.1 KRK02283.1 KRK02284.1 KRK02285.1 KRK02286.1 KRK02287.1 GBYB01003935 GBYB01003936 JAG73702.1 JAG73703.1 GAKP01008095 GAKP01008093 GAKP01008092 JAC50857.1 JXJN01002301 CH479188 EDW40204.1 BT120009 BT120010 ACZ94121.1 ACZ94122.1 CCAG010010279 EZ423999 ADD20275.1 AE014296 AY069511 CP012525 ALC44161.1 CH963847 EDW72861.1 KRF97644.1 CH477792 EAT36164.1 CH940647 EDW70481.1 GDHF01019216 JAI33098.1 OUUW01000009 SPP84738.1 CH480826 EDW44198.1 GAPW01001491 GAPW01001490 JAC12108.1 KZ150381 PZC71090.1 KQ971311 EEZ98876.1 CM000363 EDX11218.1 GFDL01011821 JAV23224.1 DS231921 EDS26494.1 ATLV01013640 KE524876 KFB38138.1 GALA01001138 JAA93714.1 AXCP01007712 CH916366 EDV97376.1 AXCM01001493 AXCN02002423 AAAB01008981 EAA14616.2 GGFM01006519 MBW27270.1 PNF42025.1 GGFK01002342 MBW35663.1 ADMH02000009 ETN68111.1 KA646160 AFP60789.1 JXUM01092887 JXUM01092888 KQ564017 KXJ72934.1 CH379069 KRT07680.1 GFDL01003005 JAV32040.1 UFQS01000344 UFQT01000344 SSX03032.1 SSX23397.1 GAKP01008094 JAC50858.1 GBRD01016415 JAG49411.1 KRG06335.1 AGB94797.1
GDQN01009915 GDQN01002893 JAT81139.1 JAT88161.1 DQ311356 ABD36300.1 KQ460500 KPJ14203.1 KQ459589 KPI97717.1 NWSH01001904 PCG69728.1 NNAY01002185 OXU21853.1 AAZX01004642 GL437061 EFN71190.1 KK107064 QOIP01000008 EZA60920.1 RLU19693.1 KZ288212 PBC32815.1 KQ435034 KZC14052.1 KQ980765 KYN13367.1 GL888063 EGI68444.1 KQ976641 KYM78805.1 KQ981272 KYN43853.1 ADTU01006100 ADTU01006101 KK854169 PTY12492.1 GBHO01015205 GBRD01007346 GDHC01012256 GDHC01003745 JAG28399.1 JAG58475.1 JAQ06373.1 JAQ14884.1 GFTR01006260 JAW10166.1 NEVH01002556 PNF42027.1 LJIG01009252 KRT83452.1 AK417407 BAN20622.1 GDKW01000912 JAI55683.1 ACPB03022348 KK853149 KDR10692.1 GBGD01001490 JAC87399.1 GBXI01013961 JAD00331.1 GEZM01027433 JAV86743.1 GEDC01006518 JAS30780.1 GECU01030690 JAS77016.1 GALX01002780 JAB65686.1 ABLF02032620 GGMS01014179 MBY83382.1 GGMR01000826 MBY13445.1 GDAI01001516 JAI16087.1 GAMC01013811 JAB92744.1 APGK01051689 APGK01051690 APGK01051691 APGK01051692 APGK01051693 KB741201 KB632375 ENN72951.1 ERL93875.1 GEBQ01003463 JAT36514.1 GECZ01022892 GECZ01015711 GECZ01002578 JAS46877.1 JAS54058.1 JAS67191.1 BABH01040929 BABH01040930 BABH01040931 KQ982691 KYQ52216.1 CM002912 KMZ00747.1 KMZ00748.1 KMZ00749.1 KMZ00752.1 KMZ00753.1 KMZ00754.1 KMZ00755.1 CH933809 EDW18722.1 KRG06336.1 CH954178 EDV52493.1 KQS44302.1 KQS44303.1 KQS44304.1 CH902618 EDV40059.1 KPU78351.1 KPU78352.1 CM000159 EDW95192.1 KRK02283.1 KRK02284.1 KRK02285.1 KRK02286.1 KRK02287.1 GBYB01003935 GBYB01003936 JAG73702.1 JAG73703.1 GAKP01008095 GAKP01008093 GAKP01008092 JAC50857.1 JXJN01002301 CH479188 EDW40204.1 BT120009 BT120010 ACZ94121.1 ACZ94122.1 CCAG010010279 EZ423999 ADD20275.1 AE014296 AY069511 CP012525 ALC44161.1 CH963847 EDW72861.1 KRF97644.1 CH477792 EAT36164.1 CH940647 EDW70481.1 GDHF01019216 JAI33098.1 OUUW01000009 SPP84738.1 CH480826 EDW44198.1 GAPW01001491 GAPW01001490 JAC12108.1 KZ150381 PZC71090.1 KQ971311 EEZ98876.1 CM000363 EDX11218.1 GFDL01011821 JAV23224.1 DS231921 EDS26494.1 ATLV01013640 KE524876 KFB38138.1 GALA01001138 JAA93714.1 AXCP01007712 CH916366 EDV97376.1 AXCM01001493 AXCN02002423 AAAB01008981 EAA14616.2 GGFM01006519 MBW27270.1 PNF42025.1 GGFK01002342 MBW35663.1 ADMH02000009 ETN68111.1 KA646160 AFP60789.1 JXUM01092887 JXUM01092888 KQ564017 KXJ72934.1 CH379069 KRT07680.1 GFDL01003005 JAV32040.1 UFQS01000344 UFQT01000344 SSX03032.1 SSX23397.1 GAKP01008094 JAC50858.1 GBRD01016415 JAG49411.1 KRG06335.1 AGB94797.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000218220
UP000215335
UP000002358
+ More
UP000000311 UP000053097 UP000279307 UP000242457 UP000076502 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000235965 UP000015103 UP000027135 UP000007819 UP000019118 UP000030742 UP000005204 UP000075809 UP000192221 UP000009192 UP000008711 UP000007801 UP000002282 UP000092460 UP000008744 UP000092445 UP000092444 UP000000803 UP000092553 UP000007798 UP000008820 UP000008792 UP000268350 UP000001292 UP000075920 UP000007266 UP000000304 UP000002320 UP000030765 UP000075880 UP000001070 UP000075883 UP000075886 UP000075884 UP000007062 UP000075900 UP000075881 UP000069272 UP000075885 UP000076408 UP000000673 UP000095301 UP000069940 UP000249989 UP000001819
UP000000311 UP000053097 UP000279307 UP000242457 UP000076502 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000235965 UP000015103 UP000027135 UP000007819 UP000019118 UP000030742 UP000005204 UP000075809 UP000192221 UP000009192 UP000008711 UP000007801 UP000002282 UP000092460 UP000008744 UP000092445 UP000092444 UP000000803 UP000092553 UP000007798 UP000008820 UP000008792 UP000268350 UP000001292 UP000075920 UP000007266 UP000000304 UP000002320 UP000030765 UP000075880 UP000001070 UP000075883 UP000075886 UP000075884 UP000007062 UP000075900 UP000075881 UP000069272 UP000075885 UP000076408 UP000000673 UP000095301 UP000069940 UP000249989 UP000001819
Interpro
Gene 3D
ProteinModelPortal
I4DNA6
A0A3S2M1H9
S4P221
A0A1E1WMS9
Q2F5R7
A0A194R8P0
+ More
A0A194PWL8 A0A2A4JDH0 A0A232EU20 K7IXJ8 E2A5U8 A0A026WXX0 A0A2A3EM31 A0A154PQ89 A0A195DKK3 F4WBG5 A0A195B3F9 A0A195FTR3 A0A158P126 A0A2R7VYF7 A0A0A9Y8A5 A0A224XCL0 A0A2J7RMF9 A0A0T6B7W7 R4WRS9 A0A0P4VUH7 T1HR39 A0A067QX62 A0A069DZ03 A0A0A1WNW3 A0A1Y1MM32 A0A1B6DYM6 A0A1B6HQR2 V5I9K4 J9JYP0 A0A2S2R218 A0A2S2N8H0 A0A0K8TNX1 W8BUE3 N6TX29 A0A1B6MKS2 A0A1B6FV35 A0A158P127 H9JXY0 A0A151WWM3 A0A1W4W6E5 A0A0J9RYL4 B4KY20 B3NIJ7 B3M975 B4PFA6 A0A0C9R7G1 A0A034W963 A0A1B0ARB1 B4GR19 A0A1A9ZFX9 D1Z373 D3TRE7 Q9VPH7 A0A0M5J8C5 B4MLK5 Q16P70 B4LGF2 A0A0K8V2R2 A0A3B0KLP6 B4IA85 A0A182W7Y2 A0A023ES91 A0A2W1BDZ4 D6WCG2 B4QRX2 A0A1Q3F6M8 B0WFV9 A0A084VJJ4 T1D417 A0A182JB29 B4IWX5 A0A182ML94 A0A182QFY0 A0A182NP75 Q7PLZ0 A0A182RS95 A0A182KE76 A0A182FLW9 A0A182PWU4 A0A182YDX7 A0A2M3ZFR8 A0A2J7RMG1 A0A2M4A4E1 W5JVA6 T1PDF9 A0A182GW91 A0A0R3P821 A0A1Q3FWQ6 A0A336KGH0 A0A034WAN3 A0A0K8S805 A0A1W4VUF2 A0A0Q9XCM5 M9PD27
A0A194PWL8 A0A2A4JDH0 A0A232EU20 K7IXJ8 E2A5U8 A0A026WXX0 A0A2A3EM31 A0A154PQ89 A0A195DKK3 F4WBG5 A0A195B3F9 A0A195FTR3 A0A158P126 A0A2R7VYF7 A0A0A9Y8A5 A0A224XCL0 A0A2J7RMF9 A0A0T6B7W7 R4WRS9 A0A0P4VUH7 T1HR39 A0A067QX62 A0A069DZ03 A0A0A1WNW3 A0A1Y1MM32 A0A1B6DYM6 A0A1B6HQR2 V5I9K4 J9JYP0 A0A2S2R218 A0A2S2N8H0 A0A0K8TNX1 W8BUE3 N6TX29 A0A1B6MKS2 A0A1B6FV35 A0A158P127 H9JXY0 A0A151WWM3 A0A1W4W6E5 A0A0J9RYL4 B4KY20 B3NIJ7 B3M975 B4PFA6 A0A0C9R7G1 A0A034W963 A0A1B0ARB1 B4GR19 A0A1A9ZFX9 D1Z373 D3TRE7 Q9VPH7 A0A0M5J8C5 B4MLK5 Q16P70 B4LGF2 A0A0K8V2R2 A0A3B0KLP6 B4IA85 A0A182W7Y2 A0A023ES91 A0A2W1BDZ4 D6WCG2 B4QRX2 A0A1Q3F6M8 B0WFV9 A0A084VJJ4 T1D417 A0A182JB29 B4IWX5 A0A182ML94 A0A182QFY0 A0A182NP75 Q7PLZ0 A0A182RS95 A0A182KE76 A0A182FLW9 A0A182PWU4 A0A182YDX7 A0A2M3ZFR8 A0A2J7RMG1 A0A2M4A4E1 W5JVA6 T1PDF9 A0A182GW91 A0A0R3P821 A0A1Q3FWQ6 A0A336KGH0 A0A034WAN3 A0A0K8S805 A0A1W4VUF2 A0A0Q9XCM5 M9PD27
PDB
3E1Y
E-value=0,
Score=1937
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
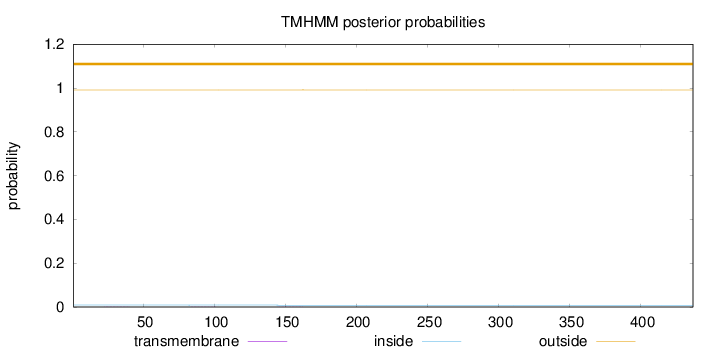
Length:
437
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0121
Exp number, first 60 AAs:
0.00372
Total prob of N-in:
0.00907
outside
1 - 437
Population Genetic Test Statistics
Pi
227.112708
Theta
191.676967
Tajima's D
0.917262
CLR
0.320861
CSRT
0.63746812659367
Interpretation
Uncertain
