Gene
KWMTBOMO14524
Annotation
PREDICTED:_uncharacterized_protein_LOC105842971_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.128
Sequence
CDS
ATGCCGCGTGTTTATAAACCTGATCCTAGAGGGAAAAGATATAGAAAGTACGATGAAAATATTATAAATGAAGCTGTTGCCGCATATGCGAATGGCAAATTCTCTTTAAAGGCCATAGCGGAGAAATATGATATTGATAAATCTGTACTTTATAGACACAGTGTCAAAACTATGAAAAAACAAGGAGGGCAAACTATTTTAACTAACGAGACTGAAGAATGCATGATAAAGTATATCAATATATGTGCAGAATGGGGATGCCCTTTAGATGCTTTAGATTTGAGATACATTGTTAAGACTTATCTTGACCAAGTGGGTCGGACAGTACTCAAATTTAAAAATAATTTACCAGGCCCCGACTTTGTCGCGAGTTTTCTAAAACGTCACAAAAACCAAATTTCCCAAAGATATAGCCAAAACATTAAGAAAAAAAGGGCAGAAGTTTCACCAAACTTGATTAAAGAATATTTCAAAGAGTTAGAAGTGTCATTGGCAGGTATTAATCCGAGTTATATAATTAATTACGATGAAACTAATTTGACCGATGACCCAGGACGCAAAAAAATCATTACTAAACGAGGAACGAAGTACCCAGAGAGGGTAATGAATAGTTCAAAGTCTTCAGTTTCATTGATGATAGCCGCTACAGCTGACGGCACTTTACTCCCGCCGTATGTGGTATACAAGGCTCAAAACTTGTATAACACATGGACTGAAAATGGACCACCAGGAGCAAGGTATAATCGTACGCAATCCGGTTGGTTTGATGCAACTATTTTTGAAGATTGGATTAAATCCATCATTATACCTCACTTCAAAGATAAAGTTGGGAAAAAGTGCCTAATAGGCGATAATCTCTCGTCGCATTTATCGATTGAGACAATAAAACTGTGCCACGAACAAAATATATCGTTCATTTTTTTACCGGCAAATAGTACACACCTCACACAACCTCTTGATGTAGCATTCTTCAGGCCTATGAAGATAGCATGGCGAAACGTAGTATTGAAGTGGAAAAAAACCGATGGGAAAGCCCAATCAACGATTCCAAAGGGATGTTTTCCTAGACTTTTAAATAAATTAATGGAAGAGCTTCGTAACAACAGTAAGGCTAATATAATAGCAGGATTCCGGAAAACAGGAATATACCCGATTAATGAAGAAGAGGTTTTAAGCAGACTACCTGAAGTGCAAAACAATAAAAAGAAAACTGAAATAGAAAAATCCGTTCTAGATTTGCTCAGGGAGATGAGATATGGAACCATGAACATAACCGAACCAAAAAGGAAAAAAAAGTTGATAGTAATACCTGGACGAAGCGTTGGTTCTGAAAGTGAAGACTACATAGAAACAGAACAAGAAGAAAGCCAACAAAAACCAAAAAAGGCAGATCTGAGATCCAAAATAACACAAGACAAAGAAAATACTACAACTAATACTTACGATGGCAAAGAAAACAATAAATATAAGGGAAAAGGTGTAGGGAAAAAAACTAAAACACAAGATCAGAATATTGCTGAAAAAGAAAATAAACAAGAAAACGGACCAGACACTGAAAAAATACTACTTGACGCAATAAATGACTTTGATTTTGTAGCGAACAATTCAATAGAAACAATGCCTATTATGTTGGAAGACATGTTGCTGTGTGAAAATGAAATTGTTTTTCATTACATACAAATAAATGAGATGCAGAATGATACCGGAATAGAAATGACGTATAAACAAAAAAATTCACCGGAAAAAAATAAAAGAAAACTGAAAATTATATCTAATGAAAAGATATCAGATATTACTACGATGAAAAAGGTTACTTTGAATAAGTGCAAAAAAATATATCCCAATTCGCGTCTTAAAATTTTAAAGGATACAATAATTGATAATGTTAATCGTCCTGGTCCAAGCAGTTTACAAACTATCCAAACTCAGCGACCATCGTTTTACAAAAACGATAATGATATTTTGAAAGATTTAATGGATGACGATATATAA
Protein
MPRVYKPDPRGKRYRKYDENIINEAVAAYANGKFSLKAIAEKYDIDKSVLYRHSVKTMKKQGGQTILTNETEECMIKYINICAEWGCPLDALDLRYIVKTYLDQVGRTVLKFKNNLPGPDFVASFLKRHKNQISQRYSQNIKKKRAEVSPNLIKEYFKELEVSLAGINPSYIINYDETNLTDDPGRKKIITKRGTKYPERVMNSSKSSVSLMIAATADGTLLPPYVVYKAQNLYNTWTENGPPGARYNRTQSGWFDATIFEDWIKSIIIPHFKDKVGKKCLIGDNLSSHLSIETIKLCHEQNISFIFLPANSTHLTQPLDVAFFRPMKIAWRNVVLKWKKTDGKAQSTIPKGCFPRLLNKLMEELRNNSKANIIAGFRKTGIYPINEEEVLSRLPEVQNNKKKTEIEKSVLDLLREMRYGTMNITEPKRKKKLIVIPGRSVGSESEDYIETEQEESQQKPKKADLRSKITQDKENTTTNTYDGKENNKYKGKGVGKKTKTQDQNIAEKENKQENGPDTEKILLDAINDFDFVANNSIETMPIMLEDMLLCENEIVFHYIQINEMQNDTGIEMTYKQKNSPEKNKRKLKIISNEKISDITTMKKVTLNKCKKIYPNSRLKILKDTIIDNVNRPGPSSLQTIQTQRPSFYKNDNDILKDLMDDDI
Summary
Uniprot
EMBL
KQ460500
KPJ14194.1
ABLF02024787
GGMR01005791
MBY18410.1
GECZ01009826
+ More
JAS59943.1 GEBQ01027923 JAT12054.1 ABLF02031950 UFQS01000373 UFQT01000373 SSX03288.1 SSX23654.1 ABLF02014120 ABLF02031335 GEBQ01022752 JAT17225.1 ABLF02024890 ABLF02042417 GFXV01002448 MBW14253.1 ABLF02032408 ABLF02032409 ABLF02032413 GEZM01096654 GEZM01096605 JAV54775.1 ABLF02013132 ABLF02013134 ABLF02013135
JAS59943.1 GEBQ01027923 JAT12054.1 ABLF02031950 UFQS01000373 UFQT01000373 SSX03288.1 SSX23654.1 ABLF02014120 ABLF02031335 GEBQ01022752 JAT17225.1 ABLF02024890 ABLF02042417 GFXV01002448 MBW14253.1 ABLF02032408 ABLF02032409 ABLF02032413 GEZM01096654 GEZM01096605 JAV54775.1 ABLF02013132 ABLF02013134 ABLF02013135
Proteomes
Interpro
ProteinModelPortal
Ontologies
KEGG
GO
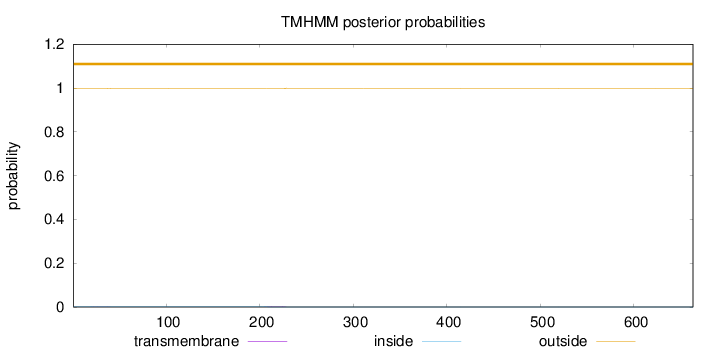
Topology
Subcellular location
Nucleus
Length:
663
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05193
Exp number, first 60 AAs:
0.01876
Total prob of N-in:
0.00294
outside
1 - 663
Population Genetic Test Statistics
Pi
324.055607
Theta
189.142865
Tajima's D
2.2485
CLR
17.730492
CSRT
0.915904204789761
Interpretation
Uncertain
