Gene
KWMTBOMO14520
Pre Gene Modal
BGIBMGA014382
Annotation
PREDICTED:_uncharacterized_protein_LOC105387121_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 1.326 Mitochondrial Reliability : 1.791 Nuclear Reliability : 1.249
Sequence
CDS
ATGGCCTCATTACCAAAATGTCGTTTAGAGTCCGATATTTATCCCTTCCAACATACTGTCGGTGTTGATATAGGAGACGAATATCATTATAATTTGATCGTCGCAAAGCTTAGTAGAGATAATGTGGAACAGATCAGTGATTTAGGCCTGACACGCCCAGACTCCCAGAAATACGAGGCATTGAAGACCAGATTGCTTGCCGTTTACGAGGAATCCGAGGTTCGGCAAGTACCAAAATTCCTGAATGAAATGGAACTGGGTGATCAAAAGCCACCCCAGCTTCTACGAAGAATGAAGGTTCTTGCCAGAAATAAATTCGCGGAAGAAACTCTTCGCATGCTGTGGATGGGACATTTACCCGTCGGCAATACAGTTATCCGTCAGTGA
Protein
MASLPKCRLESDIYPFQHTVGVDIGDEYHYNLIVAKLSRDNVEQISDLGLTRPDSQKYEALKTRLLAVYEESEVRQVPKFLNEMELGDQKPPQLLRRMKVLARNKFAEETLRMLWMGHLPVGNTVIRQ
Summary
Uniprot
H9JXW4
A0A2A4JQB6
A0A2A4JNQ1
H9J4N2
H9JCU7
A0A2H1WB17
+ More
A0A2H1V5N9 A0A0L7LSS0 A0A0L7KZ22 A0A0L7LKK8 A0A2A4JI65 A0A0L7LQI4 A0A1E1WLV2 A0A2A4JUK3 A0A2W1BAJ4 A0A0L7LG32 A0A0L7L5A2 A0A0L7LFW5 A0A2W1BA89 A0A0L7KTE7 A0A0L7KS03 A0A0J7K6V1 A0A151IIJ9 A0A1Y1MA44 A0A0A9Y7A4 A0A087TXF8 A0A026VRX8 A0A2L2YSA7 T1IE85 A0A1Y1MVP9 A0A151I8D5 A0A139W8G7 A0A139WA14 A0A0A9Z2L8 X1X8P9 K7K066 K7JBC2 A0A139WPP8 E2BRX8 A0A139WPG8 A0A0L7LFF1 A0A1I8Q981 X1WYV7 D7GYF3 D7EJX5 A0A2A4J536 A0A0A9YNN7 A0A0A9ZC79 K7JBB9 K7JBC0 K7JVT0 A0A0P8Y298 A0A0P8Y4V5 D6WTN9 T1IE13 T1I831 A0A034V7S3 A0A154PT26 A0A151JR52 A0A0P8ZL89 A0A0A9X9N4 T1HGU2 A0A139W9J0 A0A151ITG2 A0A151IZ47 A0A2M4AQA2 A0A2M4AQR5 A0A2M4ARB7 A0A2M4AYW0 A0A2M4AYL6 A0A1S4EQ14 A0A151J3D2 A0A2R7W7C8 E2BBH0 K7JBZ8 K7JX19 A0A0A9ZFJ2 A0A151JX20 A0A0P8Y533 A0A0K8SFH9 A0A151K218 A0A3Q0JNT2 A0A2M4AUN5 X1WIV3 A0A0L7QK36 A0A2M4ATJ7 A0A0L7R2Y5 A0A0J7K6I6 A0A1I8Q298 A0A151JF00 A0A151JCH0 A0A2A4J8M7 A0A0J7K0A7
A0A2H1V5N9 A0A0L7LSS0 A0A0L7KZ22 A0A0L7LKK8 A0A2A4JI65 A0A0L7LQI4 A0A1E1WLV2 A0A2A4JUK3 A0A2W1BAJ4 A0A0L7LG32 A0A0L7L5A2 A0A0L7LFW5 A0A2W1BA89 A0A0L7KTE7 A0A0L7KS03 A0A0J7K6V1 A0A151IIJ9 A0A1Y1MA44 A0A0A9Y7A4 A0A087TXF8 A0A026VRX8 A0A2L2YSA7 T1IE85 A0A1Y1MVP9 A0A151I8D5 A0A139W8G7 A0A139WA14 A0A0A9Z2L8 X1X8P9 K7K066 K7JBC2 A0A139WPP8 E2BRX8 A0A139WPG8 A0A0L7LFF1 A0A1I8Q981 X1WYV7 D7GYF3 D7EJX5 A0A2A4J536 A0A0A9YNN7 A0A0A9ZC79 K7JBB9 K7JBC0 K7JVT0 A0A0P8Y298 A0A0P8Y4V5 D6WTN9 T1IE13 T1I831 A0A034V7S3 A0A154PT26 A0A151JR52 A0A0P8ZL89 A0A0A9X9N4 T1HGU2 A0A139W9J0 A0A151ITG2 A0A151IZ47 A0A2M4AQA2 A0A2M4AQR5 A0A2M4ARB7 A0A2M4AYW0 A0A2M4AYL6 A0A1S4EQ14 A0A151J3D2 A0A2R7W7C8 E2BBH0 K7JBZ8 K7JX19 A0A0A9ZFJ2 A0A151JX20 A0A0P8Y533 A0A0K8SFH9 A0A151K218 A0A3Q0JNT2 A0A2M4AUN5 X1WIV3 A0A0L7QK36 A0A2M4ATJ7 A0A0L7R2Y5 A0A0J7K6I6 A0A1I8Q298 A0A151JF00 A0A151JCH0 A0A2A4J8M7 A0A0J7K0A7
Pubmed
EMBL
BABH01040601
NWSH01000903
PCG73612.1
PCG73611.1
BABH01022003
BABH01027731
+ More
BABH01027732 ODYU01007352 SOQ50042.1 ODYU01000815 SOQ36165.1 JTDY01000162 KOB78510.1 JTDY01004351 KOB68269.1 JTDY01000727 KOB76088.1 NWSH01001424 PCG71274.1 JTDY01000325 KOB77690.1 GDQN01003070 JAT87984.1 NWSH01000589 PCG75479.1 KZ150191 PZC72412.1 JTDY01001264 KOB74344.1 JTDY01002934 KOB70464.1 JTDY01001300 KOB74269.1 KZ150227 PZC72038.1 JTDY01006055 KOB66331.1 JTDY01006677 KOB65789.1 LBMM01012923 LBMM01012917 KMQ85926.1 KMQ85930.1 KQ977516 KYN02153.1 GEZM01036621 JAV82564.1 GBHO01016636 JAG26968.1 KK117196 KFM69797.1 KK111495 EZA46390.1 IAAA01053498 IAAA01053499 LAA11042.1 ACPB03034542 GEZM01019580 GEZM01019578 JAV89619.1 KQ978367 KYM94586.1 KQ973343 KXZ75568.1 KQ971745 KYB24749.1 GBHO01005513 JAG38091.1 ABLF02062577 AAZX01005161 AAZX01000249 KQ971307 KYB29887.1 GL450083 EFN81554.1 KYB29888.1 KOB74268.1 ABLF02062578 GG695519 EFA13383.1 KQ971759 EFA12902.2 NWSH01003151 PCG66856.1 GBHO01008962 JAG34642.1 GBHO01004189 JAG39415.1 AAZX01023342 AAZX01004730 AAZX01008012 CH902689 KPU72637.1 CH902694 KPU81747.1 KQ971355 EFA07205.2 ACPB03037918 ACPB03012735 GAKP01020775 JAC38177.1 KQ435204 KZC15052.1 KQ978645 KYN29457.1 CH902629 KPU75574.1 GBHO01027248 JAG16356.1 ACPB03011651 KQ972226 KYB24577.1 KQ981018 KYN10257.1 KQ980726 KYN14011.1 GGFK01009622 MBW42943.1 GGFK01009800 MBW43121.1 GGFK01010016 MBW43337.1 GGFK01012457 MBW45778.1 GGFK01012556 MBW45877.1 KQ980296 KYN16851.1 KK854387 PTY15358.1 GL447057 EFN86960.1 AAZX01023221 GBHO01000305 JAG43299.1 KQ981607 KYN39392.1 CH905996 KPU81830.1 GBRD01013927 JAG51899.1 LKEX01010536 KYN50169.1 GGFK01011186 MBW44507.1 ABLF02032144 KQ414987 KOC58988.1 GGFK01010782 MBW44103.1 KQ414663 KOC65240.1 LBMM01013103 KMQ85816.1 KQ979024 KYN24351.1 KQ979048 KYN23218.1 NWSH01002598 PCG67884.1 LBMM01018089 KMQ83878.1
BABH01027732 ODYU01007352 SOQ50042.1 ODYU01000815 SOQ36165.1 JTDY01000162 KOB78510.1 JTDY01004351 KOB68269.1 JTDY01000727 KOB76088.1 NWSH01001424 PCG71274.1 JTDY01000325 KOB77690.1 GDQN01003070 JAT87984.1 NWSH01000589 PCG75479.1 KZ150191 PZC72412.1 JTDY01001264 KOB74344.1 JTDY01002934 KOB70464.1 JTDY01001300 KOB74269.1 KZ150227 PZC72038.1 JTDY01006055 KOB66331.1 JTDY01006677 KOB65789.1 LBMM01012923 LBMM01012917 KMQ85926.1 KMQ85930.1 KQ977516 KYN02153.1 GEZM01036621 JAV82564.1 GBHO01016636 JAG26968.1 KK117196 KFM69797.1 KK111495 EZA46390.1 IAAA01053498 IAAA01053499 LAA11042.1 ACPB03034542 GEZM01019580 GEZM01019578 JAV89619.1 KQ978367 KYM94586.1 KQ973343 KXZ75568.1 KQ971745 KYB24749.1 GBHO01005513 JAG38091.1 ABLF02062577 AAZX01005161 AAZX01000249 KQ971307 KYB29887.1 GL450083 EFN81554.1 KYB29888.1 KOB74268.1 ABLF02062578 GG695519 EFA13383.1 KQ971759 EFA12902.2 NWSH01003151 PCG66856.1 GBHO01008962 JAG34642.1 GBHO01004189 JAG39415.1 AAZX01023342 AAZX01004730 AAZX01008012 CH902689 KPU72637.1 CH902694 KPU81747.1 KQ971355 EFA07205.2 ACPB03037918 ACPB03012735 GAKP01020775 JAC38177.1 KQ435204 KZC15052.1 KQ978645 KYN29457.1 CH902629 KPU75574.1 GBHO01027248 JAG16356.1 ACPB03011651 KQ972226 KYB24577.1 KQ981018 KYN10257.1 KQ980726 KYN14011.1 GGFK01009622 MBW42943.1 GGFK01009800 MBW43121.1 GGFK01010016 MBW43337.1 GGFK01012457 MBW45778.1 GGFK01012556 MBW45877.1 KQ980296 KYN16851.1 KK854387 PTY15358.1 GL447057 EFN86960.1 AAZX01023221 GBHO01000305 JAG43299.1 KQ981607 KYN39392.1 CH905996 KPU81830.1 GBRD01013927 JAG51899.1 LKEX01010536 KYN50169.1 GGFK01011186 MBW44507.1 ABLF02032144 KQ414987 KOC58988.1 GGFK01010782 MBW44103.1 KQ414663 KOC65240.1 LBMM01013103 KMQ85816.1 KQ979024 KYN24351.1 KQ979048 KYN23218.1 NWSH01002598 PCG67884.1 LBMM01018089 KMQ83878.1
Proteomes
Pfam
Interpro
IPR013057
AA_transpt_TM
+ More
IPR021109 Peptidase_aspartic_dom_sf
IPR018289 MULE_transposase_dom
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR034132 RP_Saci-like
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR036397 RNaseH_sf
IPR041373 RT_RNaseH
IPR041577 RT_RNaseH_2
IPR018061 Retropepsins
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR021109 Peptidase_aspartic_dom_sf
IPR018289 MULE_transposase_dom
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR034132 RP_Saci-like
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR036397 RNaseH_sf
IPR041373 RT_RNaseH
IPR041577 RT_RNaseH_2
IPR018061 Retropepsins
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
Gene 3D
ProteinModelPortal
H9JXW4
A0A2A4JQB6
A0A2A4JNQ1
H9J4N2
H9JCU7
A0A2H1WB17
+ More
A0A2H1V5N9 A0A0L7LSS0 A0A0L7KZ22 A0A0L7LKK8 A0A2A4JI65 A0A0L7LQI4 A0A1E1WLV2 A0A2A4JUK3 A0A2W1BAJ4 A0A0L7LG32 A0A0L7L5A2 A0A0L7LFW5 A0A2W1BA89 A0A0L7KTE7 A0A0L7KS03 A0A0J7K6V1 A0A151IIJ9 A0A1Y1MA44 A0A0A9Y7A4 A0A087TXF8 A0A026VRX8 A0A2L2YSA7 T1IE85 A0A1Y1MVP9 A0A151I8D5 A0A139W8G7 A0A139WA14 A0A0A9Z2L8 X1X8P9 K7K066 K7JBC2 A0A139WPP8 E2BRX8 A0A139WPG8 A0A0L7LFF1 A0A1I8Q981 X1WYV7 D7GYF3 D7EJX5 A0A2A4J536 A0A0A9YNN7 A0A0A9ZC79 K7JBB9 K7JBC0 K7JVT0 A0A0P8Y298 A0A0P8Y4V5 D6WTN9 T1IE13 T1I831 A0A034V7S3 A0A154PT26 A0A151JR52 A0A0P8ZL89 A0A0A9X9N4 T1HGU2 A0A139W9J0 A0A151ITG2 A0A151IZ47 A0A2M4AQA2 A0A2M4AQR5 A0A2M4ARB7 A0A2M4AYW0 A0A2M4AYL6 A0A1S4EQ14 A0A151J3D2 A0A2R7W7C8 E2BBH0 K7JBZ8 K7JX19 A0A0A9ZFJ2 A0A151JX20 A0A0P8Y533 A0A0K8SFH9 A0A151K218 A0A3Q0JNT2 A0A2M4AUN5 X1WIV3 A0A0L7QK36 A0A2M4ATJ7 A0A0L7R2Y5 A0A0J7K6I6 A0A1I8Q298 A0A151JF00 A0A151JCH0 A0A2A4J8M7 A0A0J7K0A7
A0A2H1V5N9 A0A0L7LSS0 A0A0L7KZ22 A0A0L7LKK8 A0A2A4JI65 A0A0L7LQI4 A0A1E1WLV2 A0A2A4JUK3 A0A2W1BAJ4 A0A0L7LG32 A0A0L7L5A2 A0A0L7LFW5 A0A2W1BA89 A0A0L7KTE7 A0A0L7KS03 A0A0J7K6V1 A0A151IIJ9 A0A1Y1MA44 A0A0A9Y7A4 A0A087TXF8 A0A026VRX8 A0A2L2YSA7 T1IE85 A0A1Y1MVP9 A0A151I8D5 A0A139W8G7 A0A139WA14 A0A0A9Z2L8 X1X8P9 K7K066 K7JBC2 A0A139WPP8 E2BRX8 A0A139WPG8 A0A0L7LFF1 A0A1I8Q981 X1WYV7 D7GYF3 D7EJX5 A0A2A4J536 A0A0A9YNN7 A0A0A9ZC79 K7JBB9 K7JBC0 K7JVT0 A0A0P8Y298 A0A0P8Y4V5 D6WTN9 T1IE13 T1I831 A0A034V7S3 A0A154PT26 A0A151JR52 A0A0P8ZL89 A0A0A9X9N4 T1HGU2 A0A139W9J0 A0A151ITG2 A0A151IZ47 A0A2M4AQA2 A0A2M4AQR5 A0A2M4ARB7 A0A2M4AYW0 A0A2M4AYL6 A0A1S4EQ14 A0A151J3D2 A0A2R7W7C8 E2BBH0 K7JBZ8 K7JX19 A0A0A9ZFJ2 A0A151JX20 A0A0P8Y533 A0A0K8SFH9 A0A151K218 A0A3Q0JNT2 A0A2M4AUN5 X1WIV3 A0A0L7QK36 A0A2M4ATJ7 A0A0L7R2Y5 A0A0J7K6I6 A0A1I8Q298 A0A151JF00 A0A151JCH0 A0A2A4J8M7 A0A0J7K0A7
Ontologies
KEGG
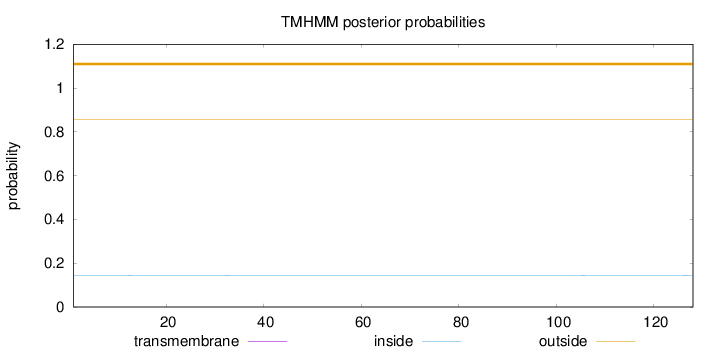
Topology
Length:
128
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00365
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.14467
outside
1 - 128
Population Genetic Test Statistics
Pi
227.339448
Theta
198.271454
Tajima's D
1.130836
CLR
24.877164
CSRT
0.702114894255287
Interpretation
Uncertain
