Gene
KWMTBOMO14516
Annotation
PREDICTED:_zinc_finger_BED_domain-containing_protein_1-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.498
Sequence
CDS
ATGCAAAGATTTCAAAAACCATTGTCTCACGTTCTTGCTGATTCTACCGACACTGAGTCTATATCTGGTACTGAATGGTCTAAAGTCGATTGCTTACTGAAAGTTATGAAGCCGTTATATGATGCTACGCAAATATCATGTGGAGTTAAATATCTTACTTTGTCAATGGTAAAACCCTTACTGTTTGGGATAATAAACGGTCTGAATAGTATCGAATTTGACTACGACGATTCATCGGAAAGGGGACAAGAATTTACAGATAATATTTTGAATGTTCTAGAAGTCAGATTTTGTGAAGTTGATCGTGATATTATTTTTAAGAAGAGTACTTATCTGGATCCCCGATACGAAAATTATTTTTATAAAGAAGAAGAAGTTAAACAAATAGAAGAAGATATTAAGCTCGAAATCGTGCGATTCCTGATGCCGGGTGATGATTCTCAAGAACTTAATGTTTTGACCAGAGAACCAAGTGGAAATCAGTATACTGGATTTTGGAGTTTCATGAAAAAACCAACAGTTAACGCCCGAGAAAAAAACCTAGAGACTCTGCGTAATGAAATATCGCAGGAAATAGTGGTGTATTTGAACTGTGGTTTAATAAGTCCTCAACAAGATCCACTTGTGGCATCTACCTACCCACCTACACACACAAACACACAATAG
Protein
MQRFQKPLSHVLADSTDTESISGTEWSKVDCLLKVMKPLYDATQISCGVKYLTLSMVKPLLFGIINGLNSIEFDYDDSSERGQEFTDNILNVLEVRFCEVDRDIIFKKSTYLDPRYENYFYKEEEVKQIEEDIKLEIVRFLMPGDDSQELNVLTREPSGNQYTGFWSFMKKPTVNAREKNLETLRNEISQEIVVYLNCGLISPQQDPLVASTYPPTHTNTQ
Summary
Uniprot
A0A088AAP1
A0A146UVL0
A0A2A3ENZ9
A0A146ZI29
A0A146QW25
G3N6T0
+ More
G3PB71 F7EC39 A0A147AD49 A0A146SGR0 J9LBU0 A0A1A8MNK0 A0A2N5VRL8 A0A146ZNJ7 J9KI18 A0A0J7KFP1 Q8T648 A0A147BCT7 A0A3P8YM71 A0A3Q3QVA5 A0A1A8N7N7 A0A2N5V0W0 A0A1S3RQ25 A0A1S3RPV5 A0A1S3QQ03 A0A3Q1FT65 A0A3N0XY52 A0A3B4WU78 A0A1A8GZ86 A0A0P8Y124 A0A1A7ZB88 A0A3N0Y7W7 A0A1S3QRY2 A0A1S3MRB9 A0A1S3QSF5 A0A1S3MR33 A0A1A8G392 A0A2N5SN73 A0A1S3QEK1
G3PB71 F7EC39 A0A147AD49 A0A146SGR0 J9LBU0 A0A1A8MNK0 A0A2N5VRL8 A0A146ZNJ7 J9KI18 A0A0J7KFP1 Q8T648 A0A147BCT7 A0A3P8YM71 A0A3Q3QVA5 A0A1A8N7N7 A0A2N5V0W0 A0A1S3RQ25 A0A1S3RPV5 A0A1S3QQ03 A0A3Q1FT65 A0A3N0XY52 A0A3B4WU78 A0A1A8GZ86 A0A0P8Y124 A0A1A7ZB88 A0A3N0Y7W7 A0A1S3QRY2 A0A1S3MRB9 A0A1S3QSF5 A0A1S3MR33 A0A1A8G392 A0A2N5SN73 A0A1S3QEK1
EMBL
GCES01077298
JAR09025.1
KZ288210
PBC32926.1
GCES01020293
JAR66030.1
+ More
GCES01126376 JAQ59946.1 AAMC01021912 GCES01009882 JAR76441.1 GCES01106986 JAQ79336.1 ABLF02031820 HAEF01016987 SBR58146.1 PGCJ01000078 PLW52612.1 GCES01018412 JAR67911.1 ABLF02036739 LBMM01008172 KMQ89087.1 AF486809 AAL93203.1 GEGO01006814 JAR88590.1 HAEH01000427 SBR64867.1 PGCJ01000145 PLW43641.1 RJVU01057296 ROK30197.1 HAEC01007697 SBQ75835.1 CH902625 KPU75502.1 HADY01001389 HAEJ01014371 SBP39874.1 RJVU01049825 ROL42306.1 HAEB01018984 SBQ65511.1 PGCJ01000915 PLW14685.1
GCES01126376 JAQ59946.1 AAMC01021912 GCES01009882 JAR76441.1 GCES01106986 JAQ79336.1 ABLF02031820 HAEF01016987 SBR58146.1 PGCJ01000078 PLW52612.1 GCES01018412 JAR67911.1 ABLF02036739 LBMM01008172 KMQ89087.1 AF486809 AAL93203.1 GEGO01006814 JAR88590.1 HAEH01000427 SBR64867.1 PGCJ01000145 PLW43641.1 RJVU01057296 ROK30197.1 HAEC01007697 SBQ75835.1 CH902625 KPU75502.1 HADY01001389 HAEJ01014371 SBP39874.1 RJVU01049825 ROL42306.1 HAEB01018984 SBQ65511.1 PGCJ01000915 PLW14685.1
Proteomes
Interpro
ProteinModelPortal
A0A088AAP1
A0A146UVL0
A0A2A3ENZ9
A0A146ZI29
A0A146QW25
G3N6T0
+ More
G3PB71 F7EC39 A0A147AD49 A0A146SGR0 J9LBU0 A0A1A8MNK0 A0A2N5VRL8 A0A146ZNJ7 J9KI18 A0A0J7KFP1 Q8T648 A0A147BCT7 A0A3P8YM71 A0A3Q3QVA5 A0A1A8N7N7 A0A2N5V0W0 A0A1S3RQ25 A0A1S3RPV5 A0A1S3QQ03 A0A3Q1FT65 A0A3N0XY52 A0A3B4WU78 A0A1A8GZ86 A0A0P8Y124 A0A1A7ZB88 A0A3N0Y7W7 A0A1S3QRY2 A0A1S3MRB9 A0A1S3QSF5 A0A1S3MR33 A0A1A8G392 A0A2N5SN73 A0A1S3QEK1
G3PB71 F7EC39 A0A147AD49 A0A146SGR0 J9LBU0 A0A1A8MNK0 A0A2N5VRL8 A0A146ZNJ7 J9KI18 A0A0J7KFP1 Q8T648 A0A147BCT7 A0A3P8YM71 A0A3Q3QVA5 A0A1A8N7N7 A0A2N5V0W0 A0A1S3RQ25 A0A1S3RPV5 A0A1S3QQ03 A0A3Q1FT65 A0A3N0XY52 A0A3B4WU78 A0A1A8GZ86 A0A0P8Y124 A0A1A7ZB88 A0A3N0Y7W7 A0A1S3QRY2 A0A1S3MRB9 A0A1S3QSF5 A0A1S3MR33 A0A1A8G392 A0A2N5SN73 A0A1S3QEK1
Ontologies
GO
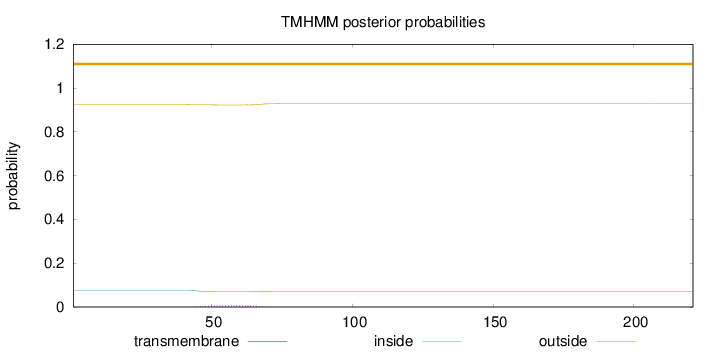
Topology
Length:
221
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16904
Exp number, first 60 AAs:
0.11106
Total prob of N-in:
0.07551
outside
1 - 221
Population Genetic Test Statistics
Pi
147.138957
Theta
115.058731
Tajima's D
0.865964
CLR
0
CSRT
0.619369031548423
Interpretation
Uncertain
