Gene
KWMTBOMO14511
Annotation
Putative_115_kDa_protein_in_type-1_retrotransposable_element_R1DM-like_Protein_[Tribolium_castaneum]
Location in the cell
Cytoplasmic Reliability : 1.689 Mitochondrial Reliability : 1.554 Nuclear Reliability : 1.382
Sequence
CDS
ATGAGAGAAGAAGAAATATCTCGTACGAAGACAGAATGGGCACAAGACTGGCAGAAGAAGGGAAAAAGTACGTGGACCCGAAAAATCATACCCAATCTTAGACCCTGGATTGAAAGAAAACATGGAGAAATAGACAGATTACTAACACAAGCTCTAAGTGGACATGGAGTCTTCAACACCTATCTCCATGCAACAGGGAAAGCAAAAAATGACATATGCCTATACTGCGAACACGTAGACACACCAGAGCACGCCCTGTTCGAATGCACAAACTTCACAGAGGTAAGAGAGAAAACGGAAGGAAACGATGGAAAAATGACAGCGGAAAACGTGATGAGAAGAATGACGAAAAGTAAATCAGGATGGGATAAATATGCGGAAATGCTCACGGAAATAGGAAAAGGTAAGGGAAAAAGACAAAAGCCTCTCTGA
Protein
MREEEISRTKTEWAQDWQKKGKSTWTRKIIPNLRPWIERKHGEIDRLLTQALSGHGVFNTYLHATGKAKNDICLYCEHVDTPEHALFECTNFTEVREKTEGNDGKMTAENVMRRMTKSKSGWDKYAEMLTEIGKGKGKRQKPL
Summary
Uniprot
A0A1B6HLL2
D7GY87
W8ANK7
W8ADE4
D7EL82
V5IAU0
+ More
U4UM48 A0A023EQZ1 U4URZ0 A0A2M4AVM9 A0A2M4AVI8 A0A2M4AU98 A0A1W7R6C5 A0A2M4AYV1 D6WC19 A0A224XIZ2 F7IYU8 A0A2S2NBZ8 A0A2S2NS54 A0A2S2P6S8 A0A139W940 F7IYV2 A0A2M4AW08 A0A1Y1KZ52 T1DI88 A0A2S2PTV0 A0A2S2NME4 A0A1W7R6I9 A0A1B6J232 A0A023EW96 A0A139W939 V5I881 A0A139W9B3 A0A139WNC9 A0A023EPM7 T1DG89 A0A139WN93 A0A139W8T3 A0A2M4BC18 K7JLB7 T1DCM0 J9LBU2 A0A2S2P2F0 A0A1W7R645 A0A2M4BC24 V5I8F3 A0A1B6HU71 A0A146MAT2 T1DG59 A0A139W9A6 A0A023EQT4 A0A0Q9WRU0 V5I7P7 A0A2M4CL45 A0A224XSH6 A0A023EPW0 A0A2S2P557 A0A2M4CSA9 K7JVH2 X1WPJ7 V5GJ14 J9LAC1 A0A2S2PIH2 A0A2M4CKQ7 A0A023EL96 A0A2M4ADW9 N6UJZ6 A0A224XA82 A0A139WAH4 U4UU36 N6UHV7 D7GXX9 A0A142LX37 J9KT70 A0A142LX43 K7JW78 A0A182GDR4 A0A0E4C7C1 A0A2S2NUP1 A0A182MNQ3 K7JP42 X1WT13 A0A2S2P0S7 U4U5X6 A0A2M4BDI3 A0A2M4BBV9 A0A2M4BC01 J9JWJ3 A0A142LX45 A0A2S2PFB9 A0A023ENU2 K7JVV4 A0A2S2Q1A1 T1I5J5 Q868R6 A0A0K8VPH1
U4UM48 A0A023EQZ1 U4URZ0 A0A2M4AVM9 A0A2M4AVI8 A0A2M4AU98 A0A1W7R6C5 A0A2M4AYV1 D6WC19 A0A224XIZ2 F7IYU8 A0A2S2NBZ8 A0A2S2NS54 A0A2S2P6S8 A0A139W940 F7IYV2 A0A2M4AW08 A0A1Y1KZ52 T1DI88 A0A2S2PTV0 A0A2S2NME4 A0A1W7R6I9 A0A1B6J232 A0A023EW96 A0A139W939 V5I881 A0A139W9B3 A0A139WNC9 A0A023EPM7 T1DG89 A0A139WN93 A0A139W8T3 A0A2M4BC18 K7JLB7 T1DCM0 J9LBU2 A0A2S2P2F0 A0A1W7R645 A0A2M4BC24 V5I8F3 A0A1B6HU71 A0A146MAT2 T1DG59 A0A139W9A6 A0A023EQT4 A0A0Q9WRU0 V5I7P7 A0A2M4CL45 A0A224XSH6 A0A023EPW0 A0A2S2P557 A0A2M4CSA9 K7JVH2 X1WPJ7 V5GJ14 J9LAC1 A0A2S2PIH2 A0A2M4CKQ7 A0A023EL96 A0A2M4ADW9 N6UJZ6 A0A224XA82 A0A139WAH4 U4UU36 N6UHV7 D7GXX9 A0A142LX37 J9KT70 A0A142LX43 K7JW78 A0A182GDR4 A0A0E4C7C1 A0A2S2NUP1 A0A182MNQ3 K7JP42 X1WT13 A0A2S2P0S7 U4U5X6 A0A2M4BDI3 A0A2M4BBV9 A0A2M4BC01 J9JWJ3 A0A142LX45 A0A2S2PFB9 A0A023ENU2 K7JVV4 A0A2S2Q1A1 T1I5J5 Q868R6 A0A0K8VPH1
Pubmed
EMBL
GECU01032129
JAS75577.1
KQ971864
EFA13315.2
GAMC01020517
JAB86038.1
+ More
GAMC01020519 GAMC01020515 JAB86040.1 DS497819 EFA11934.1 GALX01000600 JAB67866.1 KB632274 ERL91080.1 GAPW01002043 JAC11555.1 KB632419 ERL95303.1 GGFK01011357 MBW44678.1 GGFK01011482 MBW44803.1 GGFK01011045 MBW44366.1 GEHC01000964 JAV46681.1 GGFK01012648 MBW45969.1 KQ971309 EEZ99146.1 GFTR01008427 JAW07999.1 AB593321 BAK38639.1 GGMR01002122 MBY14741.1 GGMR01006977 MBY19596.1 GGMR01012534 MBY25153.1 KQ972691 KXZ75806.1 AB593323 BAK38643.1 GGFK01011630 MBW44951.1 GEZM01069016 JAV66683.1 GALA01001066 JAA93786.1 GGMR01020135 MBY32754.1 GGMR01005740 MBY18359.1 GEHC01000915 JAV46730.1 GECU01014493 JAS93213.1 GAPW01000192 JAC13406.1 KXZ75807.1 GALX01005145 JAB63321.1 KQ972544 KXZ75869.1 KQ971310 KYB29500.1 GAPW01003129 JAC10469.1 GALA01001776 JAA93076.1 KYB29499.1 KQ972982 KXZ75700.1 GGFJ01001453 MBW50594.1 AAZX01008655 GALA01001777 JAA93075.1 ABLF02031810 GGMR01010769 MBY23388.1 GEHC01001045 JAV46600.1 GGFJ01001454 MBW50595.1 GALX01004785 JAB63681.1 GECU01029521 JAS78185.1 GDHC01002080 JAQ16549.1 GALA01000302 JAA94550.1 KXZ75870.1 GAPW01002123 JAC11475.1 CH963857 KRF98317.1 GALX01006065 JAB62401.1 GGFL01001777 MBW65955.1 GFTR01004944 JAW11482.1 GAPW01002021 JAC11577.1 GGMR01011974 MBY24593.1 GGFL01004046 MBW68224.1 AAZX01013013 ABLF02013380 ABLF02013382 GALX01006869 JAB61597.1 ABLF02017307 GGMR01016419 MBY29038.1 GGFL01001583 MBW65761.1 GAPW01003605 JAC09993.1 GGFK01005660 MBW38981.1 APGK01020896 KB740258 ENN80981.1 GFTR01007160 JAW09266.1 KQ971388 KYB24913.1 KB632368 ERL93670.1 APGK01025048 KB740571 ENN80206.1 KQ972160 EFA13619.2 KU543679 AMS38363.1 ABLF02010223 KU543682 AMS38369.1 AAZX01025645 JXUM01056532 KQ561914 KXJ77178.1 HACL01000300 CFW94594.1 GGMR01008270 MBY20889.1 AXCM01017384 AXCM01017385 AAZX01012143 AAZX01012194 ABLF02014378 GGMR01010199 MBY22818.1 KB631774 ERL86016.1 GGFJ01001727 MBW50868.1 GGFJ01001369 MBW50510.1 GGFJ01001370 MBW50511.1 ABLF02013358 ABLF02013361 ABLF02054869 KU543683 AMS38371.1 GGMR01015518 MBY28137.1 GAPW01002982 JAC10616.1 GGMS01002352 MBY71555.1 ACPB03015998 ACPB03015999 ACPB03016000 ACPB03016001 AB090819 BAC57914.1 GDHF01029467 GDHF01011533 JAI22847.1 JAI40781.1
GAMC01020519 GAMC01020515 JAB86040.1 DS497819 EFA11934.1 GALX01000600 JAB67866.1 KB632274 ERL91080.1 GAPW01002043 JAC11555.1 KB632419 ERL95303.1 GGFK01011357 MBW44678.1 GGFK01011482 MBW44803.1 GGFK01011045 MBW44366.1 GEHC01000964 JAV46681.1 GGFK01012648 MBW45969.1 KQ971309 EEZ99146.1 GFTR01008427 JAW07999.1 AB593321 BAK38639.1 GGMR01002122 MBY14741.1 GGMR01006977 MBY19596.1 GGMR01012534 MBY25153.1 KQ972691 KXZ75806.1 AB593323 BAK38643.1 GGFK01011630 MBW44951.1 GEZM01069016 JAV66683.1 GALA01001066 JAA93786.1 GGMR01020135 MBY32754.1 GGMR01005740 MBY18359.1 GEHC01000915 JAV46730.1 GECU01014493 JAS93213.1 GAPW01000192 JAC13406.1 KXZ75807.1 GALX01005145 JAB63321.1 KQ972544 KXZ75869.1 KQ971310 KYB29500.1 GAPW01003129 JAC10469.1 GALA01001776 JAA93076.1 KYB29499.1 KQ972982 KXZ75700.1 GGFJ01001453 MBW50594.1 AAZX01008655 GALA01001777 JAA93075.1 ABLF02031810 GGMR01010769 MBY23388.1 GEHC01001045 JAV46600.1 GGFJ01001454 MBW50595.1 GALX01004785 JAB63681.1 GECU01029521 JAS78185.1 GDHC01002080 JAQ16549.1 GALA01000302 JAA94550.1 KXZ75870.1 GAPW01002123 JAC11475.1 CH963857 KRF98317.1 GALX01006065 JAB62401.1 GGFL01001777 MBW65955.1 GFTR01004944 JAW11482.1 GAPW01002021 JAC11577.1 GGMR01011974 MBY24593.1 GGFL01004046 MBW68224.1 AAZX01013013 ABLF02013380 ABLF02013382 GALX01006869 JAB61597.1 ABLF02017307 GGMR01016419 MBY29038.1 GGFL01001583 MBW65761.1 GAPW01003605 JAC09993.1 GGFK01005660 MBW38981.1 APGK01020896 KB740258 ENN80981.1 GFTR01007160 JAW09266.1 KQ971388 KYB24913.1 KB632368 ERL93670.1 APGK01025048 KB740571 ENN80206.1 KQ972160 EFA13619.2 KU543679 AMS38363.1 ABLF02010223 KU543682 AMS38369.1 AAZX01025645 JXUM01056532 KQ561914 KXJ77178.1 HACL01000300 CFW94594.1 GGMR01008270 MBY20889.1 AXCM01017384 AXCM01017385 AAZX01012143 AAZX01012194 ABLF02014378 GGMR01010199 MBY22818.1 KB631774 ERL86016.1 GGFJ01001727 MBW50868.1 GGFJ01001369 MBW50510.1 GGFJ01001370 MBW50511.1 ABLF02013358 ABLF02013361 ABLF02054869 KU543683 AMS38371.1 GGMR01015518 MBY28137.1 GAPW01002982 JAC10616.1 GGMS01002352 MBY71555.1 ACPB03015998 ACPB03015999 ACPB03016000 ACPB03016001 AB090819 BAC57914.1 GDHF01029467 GDHF01011533 JAI22847.1 JAI40781.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A1B6HLL2
D7GY87
W8ANK7
W8ADE4
D7EL82
V5IAU0
+ More
U4UM48 A0A023EQZ1 U4URZ0 A0A2M4AVM9 A0A2M4AVI8 A0A2M4AU98 A0A1W7R6C5 A0A2M4AYV1 D6WC19 A0A224XIZ2 F7IYU8 A0A2S2NBZ8 A0A2S2NS54 A0A2S2P6S8 A0A139W940 F7IYV2 A0A2M4AW08 A0A1Y1KZ52 T1DI88 A0A2S2PTV0 A0A2S2NME4 A0A1W7R6I9 A0A1B6J232 A0A023EW96 A0A139W939 V5I881 A0A139W9B3 A0A139WNC9 A0A023EPM7 T1DG89 A0A139WN93 A0A139W8T3 A0A2M4BC18 K7JLB7 T1DCM0 J9LBU2 A0A2S2P2F0 A0A1W7R645 A0A2M4BC24 V5I8F3 A0A1B6HU71 A0A146MAT2 T1DG59 A0A139W9A6 A0A023EQT4 A0A0Q9WRU0 V5I7P7 A0A2M4CL45 A0A224XSH6 A0A023EPW0 A0A2S2P557 A0A2M4CSA9 K7JVH2 X1WPJ7 V5GJ14 J9LAC1 A0A2S2PIH2 A0A2M4CKQ7 A0A023EL96 A0A2M4ADW9 N6UJZ6 A0A224XA82 A0A139WAH4 U4UU36 N6UHV7 D7GXX9 A0A142LX37 J9KT70 A0A142LX43 K7JW78 A0A182GDR4 A0A0E4C7C1 A0A2S2NUP1 A0A182MNQ3 K7JP42 X1WT13 A0A2S2P0S7 U4U5X6 A0A2M4BDI3 A0A2M4BBV9 A0A2M4BC01 J9JWJ3 A0A142LX45 A0A2S2PFB9 A0A023ENU2 K7JVV4 A0A2S2Q1A1 T1I5J5 Q868R6 A0A0K8VPH1
U4UM48 A0A023EQZ1 U4URZ0 A0A2M4AVM9 A0A2M4AVI8 A0A2M4AU98 A0A1W7R6C5 A0A2M4AYV1 D6WC19 A0A224XIZ2 F7IYU8 A0A2S2NBZ8 A0A2S2NS54 A0A2S2P6S8 A0A139W940 F7IYV2 A0A2M4AW08 A0A1Y1KZ52 T1DI88 A0A2S2PTV0 A0A2S2NME4 A0A1W7R6I9 A0A1B6J232 A0A023EW96 A0A139W939 V5I881 A0A139W9B3 A0A139WNC9 A0A023EPM7 T1DG89 A0A139WN93 A0A139W8T3 A0A2M4BC18 K7JLB7 T1DCM0 J9LBU2 A0A2S2P2F0 A0A1W7R645 A0A2M4BC24 V5I8F3 A0A1B6HU71 A0A146MAT2 T1DG59 A0A139W9A6 A0A023EQT4 A0A0Q9WRU0 V5I7P7 A0A2M4CL45 A0A224XSH6 A0A023EPW0 A0A2S2P557 A0A2M4CSA9 K7JVH2 X1WPJ7 V5GJ14 J9LAC1 A0A2S2PIH2 A0A2M4CKQ7 A0A023EL96 A0A2M4ADW9 N6UJZ6 A0A224XA82 A0A139WAH4 U4UU36 N6UHV7 D7GXX9 A0A142LX37 J9KT70 A0A142LX43 K7JW78 A0A182GDR4 A0A0E4C7C1 A0A2S2NUP1 A0A182MNQ3 K7JP42 X1WT13 A0A2S2P0S7 U4U5X6 A0A2M4BDI3 A0A2M4BBV9 A0A2M4BC01 J9JWJ3 A0A142LX45 A0A2S2PFB9 A0A023ENU2 K7JVV4 A0A2S2Q1A1 T1I5J5 Q868R6 A0A0K8VPH1
Ontologies
KEGG
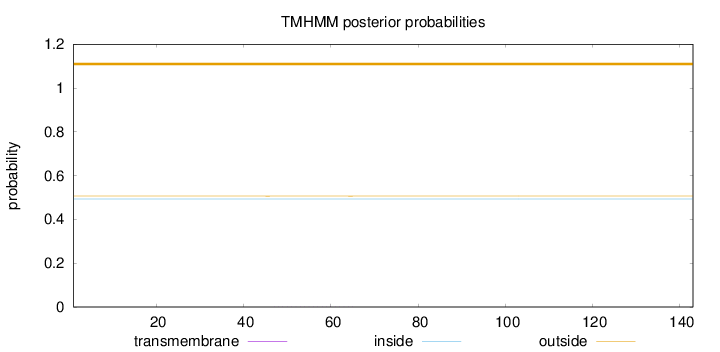
Topology
Length:
143
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00492
Exp number, first 60 AAs:
0.00369
Total prob of N-in:
0.49338
outside
1 - 143
Population Genetic Test Statistics
Pi
272.988925
Theta
208.245001
Tajima's D
0.946872
CLR
0
CSRT
0.647917604119794
Interpretation
Uncertain
