Gene
KWMTBOMO14500 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000167
Annotation
PREDICTED:_lactoylglutathione_lyase_[Papilio_polytes]
Full name
Lactoylglutathione lyase
Alternative Name
Glyoxalase I
Aldoketomutase
Ketone-aldehyde mutase
Methylglyoxalase
S-D-lactoylglutathione methylglyoxal lyase
Aldoketomutase
Ketone-aldehyde mutase
Methylglyoxalase
S-D-lactoylglutathione methylglyoxal lyase
Location in the cell
Cytoplasmic Reliability : 1.687
Sequence
CDS
ATGGCCCAAGGGATTTCTCCACAAGAAATAGAGGCTCTATGCCAGACGCCAGATCCCACTACCAAGGATTTTATGTTCCAACAAACAATGTACAGGATCAAAGATCCAAGGAAGACTATACCTTTTTACACTGGTGTCCTCGGAATGACATTACTTAAGCAACTGCATTTTCCAGCAATGAAGTTTTCGTTGTTCTTTATGGGCTATGAGAATCCAGCTGAAGTACCGAGAGACGATGACGCCAGAACTAAATGGGCCATGACTAGGAAAGCCACGTTGGAATTGACATATAACTGGGGAACAGAGAGCGACGACTCAAGCTACCACAACGGTAATTCAGATCCTCGCGGTTTCGGACATATTGGTATACTTGTGCCGGATGTGGATGTGGCGTGTGCTAGATTTGAGCAGCTAGGTGTTAAATTTATTAAACGTCCCAACGATGGTAAAATGAAAGGCCTAGCTTTCATTCAAGACCCGGACGGTTACTGGATAGAAATATTCACAAGTAATGTTGTTTGA
Protein
MAQGISPQEIEALCQTPDPTTKDFMFQQTMYRIKDPRKTIPFYTGVLGMTLLKQLHFPAMKFSLFFMGYENPAEVPRDDDARTKWAMTRKATLELTYNWGTESDDSSYHNGNSDPRGFGHIGILVPDVDVACARFEQLGVKFIKRPNDGKMKGLAFIQDPDGYWIEIFTSNVV
Summary
Description
Catalyzes the conversion of hemimercaptal, formed from methylglyoxal and glutathione, to S-lactoylglutathione.
Catalyzes the conversion of hemimercaptal, formed from methylglyoxal and glutathione, to S-lactoylglutathione. Involved in the regulation of TNF-induced transcriptional activity of NF-kappa-B (By similarity).
Catalyzes the conversion of hemimercaptal, formed from methylglyoxal and glutathione, to S-lactoylglutathione. Involved in the regulation of TNF-induced transcriptional activity of NF-kappa-B. Required for normal osteoclastogenesis.
Catalyzes the conversion of hemimercaptal, formed from methylglyoxal and glutathione, to S-lactoylglutathione. Involved in the regulation of TNF-induced transcriptional activity of NF-kappa-B (By similarity).
Catalyzes the conversion of hemimercaptal, formed from methylglyoxal and glutathione, to S-lactoylglutathione. Involved in the regulation of TNF-induced transcriptional activity of NF-kappa-B. Required for normal osteoclastogenesis.
Catalytic Activity
(R)-S-lactoylglutathione = glutathione + methylglyoxal
Cofactor
Zn(2+)
Subunit
Homodimer.
Miscellaneous
Expressed at higher levels in CD-1 mice which have been bred for low-anxiety-related behavior than in those which have been bred for high-anxiety-related behavior.
Similarity
Belongs to the glyoxalase I family.
Keywords
Acetylation
Complete proteome
Direct protein sequencing
Disulfide bond
Glutathionylation
Lyase
Metal-binding
Phosphoprotein
Reference proteome
Zinc
3D-structure
Feature
chain Lactoylglutathione lyase
Uniprot
H9ISE0
I4DJZ6
A0A0N1I7U4
S4P8F7
A0A212EK31
A0A0N0P9N5
+ More
A0A0L0CQF7 A0A1I8NAP6 A0A1I8P266 A0A1L8EER8 A0A034WRU6 A0A1B0AGD6 A0A0C9QHA5 A0A1L8E1V2 A0A0K8TTL5 A0A1A9XFG1 A0A1A9WR34 A0A1B0CWR5 A0A0K8V541 A0A0K8WLK0 W8C8F4 B3MI71 B4P1N4 A0A0A1XNB4 A0A1W4V108 B4J6N5 B4KNE1 A0A1L8E2B8 B4LMA0 U5ERW2 B3NA03 B4MR75 A0A1B0FE17 R4WQX5 A0A1B6FGH2 A0A1B6I8Z5 A0A1B6BWJ1 A0A1B0AGE6 A0A162SUJ7 A0A1S3CXY6 A1Z6X6 A0A1J1I4L4 A0A0P4W2W8 E9HSG6 A0A224XLM9 A0A0M3QUL6 A0A3B0J277 B4HQT5 B4QE88 A0A0Q3PMZ0 Q28BX7 A0A1B0G6Z7 A0A1B6KHS6 B5E0X3 B4GGM8 A0A2P8XVK5 Q5BL69 A0A2H1VUV9 A0A069DPK7 A0A1A9V386 A0A226EMY4 A0A1S4G1K4 A0A0B8RTN5 I3LDM7 Q16GF9 J9NRV6 A0A1B0AGG6 A0A1A9V3A8 K7F913 A0A1L1RZK1 A0A023F9U0 A0A182G6B7 Q6P7Q4 U3I6H4 A0A093G8X9 A0A2Y9SK07 A0A3M0KIS4 A0A1W4X1J7 R4FLZ2 A0A2I0MAE4 A5GZX3 Q9CPU0 A0A1V4JJR5 A0A098LZ64 U3K9F4 A0A340X9Z3 A0A2U4CJ44 A0A2Y9NX72 A0A2Y9JEG9 A0A091D921 F6Y7I6 A0A226PAL2 A0A0B8RRB0 A0A091JUL6 T1P962 A0A384BEX9 A0A226NE20 F7ECV7 J3SEL3 T1DKP2
A0A0L0CQF7 A0A1I8NAP6 A0A1I8P266 A0A1L8EER8 A0A034WRU6 A0A1B0AGD6 A0A0C9QHA5 A0A1L8E1V2 A0A0K8TTL5 A0A1A9XFG1 A0A1A9WR34 A0A1B0CWR5 A0A0K8V541 A0A0K8WLK0 W8C8F4 B3MI71 B4P1N4 A0A0A1XNB4 A0A1W4V108 B4J6N5 B4KNE1 A0A1L8E2B8 B4LMA0 U5ERW2 B3NA03 B4MR75 A0A1B0FE17 R4WQX5 A0A1B6FGH2 A0A1B6I8Z5 A0A1B6BWJ1 A0A1B0AGE6 A0A162SUJ7 A0A1S3CXY6 A1Z6X6 A0A1J1I4L4 A0A0P4W2W8 E9HSG6 A0A224XLM9 A0A0M3QUL6 A0A3B0J277 B4HQT5 B4QE88 A0A0Q3PMZ0 Q28BX7 A0A1B0G6Z7 A0A1B6KHS6 B5E0X3 B4GGM8 A0A2P8XVK5 Q5BL69 A0A2H1VUV9 A0A069DPK7 A0A1A9V386 A0A226EMY4 A0A1S4G1K4 A0A0B8RTN5 I3LDM7 Q16GF9 J9NRV6 A0A1B0AGG6 A0A1A9V3A8 K7F913 A0A1L1RZK1 A0A023F9U0 A0A182G6B7 Q6P7Q4 U3I6H4 A0A093G8X9 A0A2Y9SK07 A0A3M0KIS4 A0A1W4X1J7 R4FLZ2 A0A2I0MAE4 A5GZX3 Q9CPU0 A0A1V4JJR5 A0A098LZ64 U3K9F4 A0A340X9Z3 A0A2U4CJ44 A0A2Y9NX72 A0A2Y9JEG9 A0A091D921 F6Y7I6 A0A226PAL2 A0A0B8RRB0 A0A091JUL6 T1P962 A0A384BEX9 A0A226NE20 F7ECV7 J3SEL3 T1DKP2
EC Number
4.4.1.5
Pubmed
19121390
22651552
26354079
23622113
22118469
26108605
+ More
25315136 25348373 26369729 24495485 17994087 17550304 25830018 23691247 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21292972 22936249 15632085 29403074 26334808 17510324 16341006 17381049 15592404 25474469 26483478 15489334 23371554 12040188 21183079 23806337 16141072 15858064 18695250 18464734 24621616 25476704 17495919 23025625 23758969 25727380
25315136 25348373 26369729 24495485 17994087 17550304 25830018 23691247 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21292972 22936249 15632085 29403074 26334808 17510324 16341006 17381049 15592404 25474469 26483478 15489334 23371554 12040188 21183079 23806337 16141072 15858064 18695250 18464734 24621616 25476704 17495919 23025625 23758969 25727380
EMBL
BABH01024381
BABH01024382
AK401614
BAM18236.1
KQ460501
KPJ14166.1
+ More
GAIX01005956 JAA86604.1 AGBW02014345 OWR41836.1 KQ459286 KPJ02040.1 JRES01000049 KNC34608.1 GFDG01001654 JAV17145.1 GAKP01002042 JAC56910.1 GBYB01002889 JAG72656.1 GFDF01001440 JAV12644.1 GDAI01000110 JAI17493.1 AJWK01032776 GDHF01018318 JAI33996.1 GDHF01000347 JAI51967.1 GAMC01003074 JAC03482.1 CH902619 EDV35916.1 CM000157 EDW89170.1 GBXI01001947 JAD12345.1 CH916367 EDW00938.1 CH933808 EDW09994.1 GFDF01001429 JAV12655.1 CH940648 EDW60978.1 GANO01002647 JAB57224.1 CH954177 EDV59699.1 CH963849 EDW74614.1 CCAG010017792 AK418086 BAN21301.1 GECZ01020494 JAS49275.1 GECU01037144 GECU01034632 GECU01033622 GECU01024292 GECU01016033 GECU01012191 GECU01007728 JAS70562.1 JAS73074.1 JAS74084.1 JAS83414.1 JAS91673.1 JAS95515.1 JAS99978.1 GEDC01031636 GEDC01024624 JAS05662.1 JAS12674.1 LRGB01000024 KZS21640.1 AE013599 AAF59267.1 CVRI01000039 CRK94698.1 GDRN01092021 JAI60175.1 GL732751 EFX65323.1 GFTR01003049 JAW13377.1 CP012524 ALC40843.1 OUUW01000001 SPP73132.1 CH480816 EDW46745.1 CM000362 CM002911 EDX05997.1 KMY91959.1 LMAW01002759 KQK77428.1 CR942582 CAJ83987.1 CCAG010003750 GEBQ01028998 GEBQ01021307 JAT10979.1 JAT18670.1 CM000071 EDY69054.1 CH479183 EDW35648.1 PYGN01001280 PSN36047.1 BC090582 AAH90582.1 ODYU01004591 SOQ44627.1 GBGD01003129 JAC85760.1 LNIX01000002 OXA59045.1 GBZA01000248 JAG69520.1 AEMK02000055 CH478282 EAT33325.1 AAEX03008305 AGCU01109535 AGCU01109536 AGCU01109537 AADN05000005 GBBI01001024 JAC17688.1 JXUM01147679 KQ570417 KXJ68365.1 BC061570 KL215801 KFV66665.1 QRBI01000106 RMC12541.1 ACPB03022597 GAHY01001929 JAA75581.1 AKCR02000024 PKK26650.1 DQ487161 DQ487162 CH466542 ABF48483.1 ABF48484.1 EDL08651.1 AK002386 AK003567 AK005055 AK031832 AK049703 BC024663 BC081432 LSYS01007194 OPJ72431.1 GBSI01001241 JAC95255.1 AGTO01000388 KN123046 KFO26998.1 AWGT02000117 OXB76612.1 GBSH01001441 JAG67585.1 KK529555 KFP27603.1 KA645321 AFP59950.1 MCFN01000529 MCFN01000081 OXB57707.1 OXB65794.1 JU174920 GBEX01001751 AFJ50446.1 JAI12809.1 GAAZ01001233 GBKC01001843 GBKD01001113 JAA96710.1 JAG44227.1 JAG46505.1
GAIX01005956 JAA86604.1 AGBW02014345 OWR41836.1 KQ459286 KPJ02040.1 JRES01000049 KNC34608.1 GFDG01001654 JAV17145.1 GAKP01002042 JAC56910.1 GBYB01002889 JAG72656.1 GFDF01001440 JAV12644.1 GDAI01000110 JAI17493.1 AJWK01032776 GDHF01018318 JAI33996.1 GDHF01000347 JAI51967.1 GAMC01003074 JAC03482.1 CH902619 EDV35916.1 CM000157 EDW89170.1 GBXI01001947 JAD12345.1 CH916367 EDW00938.1 CH933808 EDW09994.1 GFDF01001429 JAV12655.1 CH940648 EDW60978.1 GANO01002647 JAB57224.1 CH954177 EDV59699.1 CH963849 EDW74614.1 CCAG010017792 AK418086 BAN21301.1 GECZ01020494 JAS49275.1 GECU01037144 GECU01034632 GECU01033622 GECU01024292 GECU01016033 GECU01012191 GECU01007728 JAS70562.1 JAS73074.1 JAS74084.1 JAS83414.1 JAS91673.1 JAS95515.1 JAS99978.1 GEDC01031636 GEDC01024624 JAS05662.1 JAS12674.1 LRGB01000024 KZS21640.1 AE013599 AAF59267.1 CVRI01000039 CRK94698.1 GDRN01092021 JAI60175.1 GL732751 EFX65323.1 GFTR01003049 JAW13377.1 CP012524 ALC40843.1 OUUW01000001 SPP73132.1 CH480816 EDW46745.1 CM000362 CM002911 EDX05997.1 KMY91959.1 LMAW01002759 KQK77428.1 CR942582 CAJ83987.1 CCAG010003750 GEBQ01028998 GEBQ01021307 JAT10979.1 JAT18670.1 CM000071 EDY69054.1 CH479183 EDW35648.1 PYGN01001280 PSN36047.1 BC090582 AAH90582.1 ODYU01004591 SOQ44627.1 GBGD01003129 JAC85760.1 LNIX01000002 OXA59045.1 GBZA01000248 JAG69520.1 AEMK02000055 CH478282 EAT33325.1 AAEX03008305 AGCU01109535 AGCU01109536 AGCU01109537 AADN05000005 GBBI01001024 JAC17688.1 JXUM01147679 KQ570417 KXJ68365.1 BC061570 KL215801 KFV66665.1 QRBI01000106 RMC12541.1 ACPB03022597 GAHY01001929 JAA75581.1 AKCR02000024 PKK26650.1 DQ487161 DQ487162 CH466542 ABF48483.1 ABF48484.1 EDL08651.1 AK002386 AK003567 AK005055 AK031832 AK049703 BC024663 BC081432 LSYS01007194 OPJ72431.1 GBSI01001241 JAC95255.1 AGTO01000388 KN123046 KFO26998.1 AWGT02000117 OXB76612.1 GBSH01001441 JAG67585.1 KK529555 KFP27603.1 KA645321 AFP59950.1 MCFN01000529 MCFN01000081 OXB57707.1 OXB65794.1 JU174920 GBEX01001751 AFJ50446.1 JAI12809.1 GAAZ01001233 GBKC01001843 GBKD01001113 JAA96710.1 JAG44227.1 JAG46505.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000037069
UP000095301
+ More
UP000095300 UP000092445 UP000092443 UP000091820 UP000092461 UP000007801 UP000002282 UP000192221 UP000001070 UP000009192 UP000008792 UP000008711 UP000007798 UP000092444 UP000076858 UP000079169 UP000000803 UP000183832 UP000000305 UP000092553 UP000268350 UP000001292 UP000000304 UP000051836 UP000001819 UP000008744 UP000245037 UP000078200 UP000198287 UP000008227 UP000008820 UP000002254 UP000007267 UP000000539 UP000069940 UP000249989 UP000002494 UP000053875 UP000248484 UP000269221 UP000192223 UP000015103 UP000053872 UP000000589 UP000190648 UP000016665 UP000265300 UP000245320 UP000248483 UP000248482 UP000028990 UP000002279 UP000198419 UP000261681 UP000198323 UP000002280
UP000095300 UP000092445 UP000092443 UP000091820 UP000092461 UP000007801 UP000002282 UP000192221 UP000001070 UP000009192 UP000008792 UP000008711 UP000007798 UP000092444 UP000076858 UP000079169 UP000000803 UP000183832 UP000000305 UP000092553 UP000268350 UP000001292 UP000000304 UP000051836 UP000001819 UP000008744 UP000245037 UP000078200 UP000198287 UP000008227 UP000008820 UP000002254 UP000007267 UP000000539 UP000069940 UP000249989 UP000002494 UP000053875 UP000248484 UP000269221 UP000192223 UP000015103 UP000053872 UP000000589 UP000190648 UP000016665 UP000265300 UP000245320 UP000248483 UP000248482 UP000028990 UP000002279 UP000198419 UP000261681 UP000198323 UP000002280
Pfam
PF00903 Glyoxalase
Interpro
SUPFAM
SSF54593
SSF54593
Gene 3D
ProteinModelPortal
H9ISE0
I4DJZ6
A0A0N1I7U4
S4P8F7
A0A212EK31
A0A0N0P9N5
+ More
A0A0L0CQF7 A0A1I8NAP6 A0A1I8P266 A0A1L8EER8 A0A034WRU6 A0A1B0AGD6 A0A0C9QHA5 A0A1L8E1V2 A0A0K8TTL5 A0A1A9XFG1 A0A1A9WR34 A0A1B0CWR5 A0A0K8V541 A0A0K8WLK0 W8C8F4 B3MI71 B4P1N4 A0A0A1XNB4 A0A1W4V108 B4J6N5 B4KNE1 A0A1L8E2B8 B4LMA0 U5ERW2 B3NA03 B4MR75 A0A1B0FE17 R4WQX5 A0A1B6FGH2 A0A1B6I8Z5 A0A1B6BWJ1 A0A1B0AGE6 A0A162SUJ7 A0A1S3CXY6 A1Z6X6 A0A1J1I4L4 A0A0P4W2W8 E9HSG6 A0A224XLM9 A0A0M3QUL6 A0A3B0J277 B4HQT5 B4QE88 A0A0Q3PMZ0 Q28BX7 A0A1B0G6Z7 A0A1B6KHS6 B5E0X3 B4GGM8 A0A2P8XVK5 Q5BL69 A0A2H1VUV9 A0A069DPK7 A0A1A9V386 A0A226EMY4 A0A1S4G1K4 A0A0B8RTN5 I3LDM7 Q16GF9 J9NRV6 A0A1B0AGG6 A0A1A9V3A8 K7F913 A0A1L1RZK1 A0A023F9U0 A0A182G6B7 Q6P7Q4 U3I6H4 A0A093G8X9 A0A2Y9SK07 A0A3M0KIS4 A0A1W4X1J7 R4FLZ2 A0A2I0MAE4 A5GZX3 Q9CPU0 A0A1V4JJR5 A0A098LZ64 U3K9F4 A0A340X9Z3 A0A2U4CJ44 A0A2Y9NX72 A0A2Y9JEG9 A0A091D921 F6Y7I6 A0A226PAL2 A0A0B8RRB0 A0A091JUL6 T1P962 A0A384BEX9 A0A226NE20 F7ECV7 J3SEL3 T1DKP2
A0A0L0CQF7 A0A1I8NAP6 A0A1I8P266 A0A1L8EER8 A0A034WRU6 A0A1B0AGD6 A0A0C9QHA5 A0A1L8E1V2 A0A0K8TTL5 A0A1A9XFG1 A0A1A9WR34 A0A1B0CWR5 A0A0K8V541 A0A0K8WLK0 W8C8F4 B3MI71 B4P1N4 A0A0A1XNB4 A0A1W4V108 B4J6N5 B4KNE1 A0A1L8E2B8 B4LMA0 U5ERW2 B3NA03 B4MR75 A0A1B0FE17 R4WQX5 A0A1B6FGH2 A0A1B6I8Z5 A0A1B6BWJ1 A0A1B0AGE6 A0A162SUJ7 A0A1S3CXY6 A1Z6X6 A0A1J1I4L4 A0A0P4W2W8 E9HSG6 A0A224XLM9 A0A0M3QUL6 A0A3B0J277 B4HQT5 B4QE88 A0A0Q3PMZ0 Q28BX7 A0A1B0G6Z7 A0A1B6KHS6 B5E0X3 B4GGM8 A0A2P8XVK5 Q5BL69 A0A2H1VUV9 A0A069DPK7 A0A1A9V386 A0A226EMY4 A0A1S4G1K4 A0A0B8RTN5 I3LDM7 Q16GF9 J9NRV6 A0A1B0AGG6 A0A1A9V3A8 K7F913 A0A1L1RZK1 A0A023F9U0 A0A182G6B7 Q6P7Q4 U3I6H4 A0A093G8X9 A0A2Y9SK07 A0A3M0KIS4 A0A1W4X1J7 R4FLZ2 A0A2I0MAE4 A5GZX3 Q9CPU0 A0A1V4JJR5 A0A098LZ64 U3K9F4 A0A340X9Z3 A0A2U4CJ44 A0A2Y9NX72 A0A2Y9JEG9 A0A091D921 F6Y7I6 A0A226PAL2 A0A0B8RRB0 A0A091JUL6 T1P962 A0A384BEX9 A0A226NE20 F7ECV7 J3SEL3 T1DKP2
PDB
4PV5
E-value=5.73999e-66,
Score=632
Ontologies
PATHWAY
GO
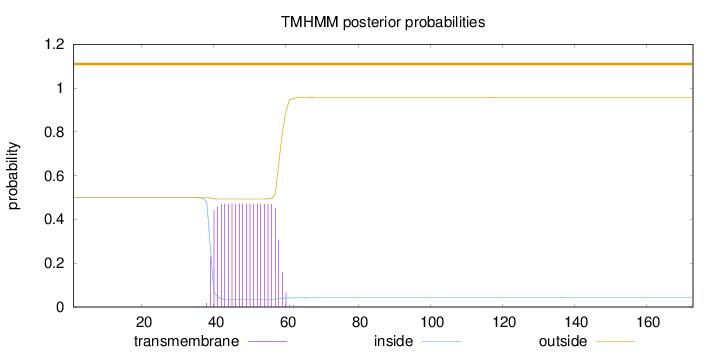
Topology
Length:
173
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.24630999999999
Exp number, first 60 AAs:
9.2117
Total prob of N-in:
0.49998
outside
1 - 173
Population Genetic Test Statistics
Pi
250.864342
Theta
167.60775
Tajima's D
1.677607
CLR
0
CSRT
0.821308934553272
Interpretation
Uncertain
