Gene
KWMTBOMO14491
Pre Gene Modal
BGIBMGA000068
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.641 PlasmaMembrane Reliability : 1.979
Sequence
CDS
ATGGCGTCGTCTTTCTTAGGAAACCTGGTGGAATCTACATATCTGCCCTTGGAAACTGCAACGACCATCCTCGCGATGGCAGGCCTTTTCAAGTGGCCCCCTCAAGCTACAGTAAATGATGGTGATCGTTTCGACTTCATCGTGATCGGGTCGGGTGTTGGAGCCGTCATCGCCAATAGATTAACAGAAAACGAAGATGTTCGAGTACTATTAATAGAAGCAGGAAAAAACCCCTCCGTCGAATCTATGCTACCCGGTCTTTTTATTCTGCTCCAAAACTCGTACCAAGATTGGAACTATGTGTCCGAACCCGAGGAAGCTACAAAAAATCAACAATTCGGTGCCTATAGAACGTCCGCAGGAAAATGTCTGGGTGGAAGCAGCAATATCAATCATTTTATACATCTACGAGGAGATCCGTGCGACTTCGATTCTTGGGCAGCCTACCTCAAAGACGAATCTTGGTCTTATAAGAACGTTCTTCCGTATTTCCGGAAAAGTGAGACAATCCAAGACGAAGACATCCTTAAATATTATGCCAATTTCCACGGTGTTGACGGTCCAGTAATTATAACCAGACAACCTGATGATTCCACCCGTAATATCATGGAGTCGTTCGAAGAAATTGGAGTACCATCGGTTTTAGACCTGAACACGAACAATACTGTTGGGTTCACGGAATCCTCTTTCATCATAGGCAACGGCCGAAGACAGAGTACTTCTCAAGCATACCTGAACAACTTGAATAGAGATAATTTGTACGTATTAACCGAAACTGTTGCTGAAAAAATTATTTTCGAAGATAATGTAGCTGTAGGTGTCATCTTAAGGCTTGGAAGCGGTGAAAAAATTACTGTCTACGCTAATAGAGAAGTCATTGTCTCTGCCGGAACATTTAACCCCCTTACCTCATCCGGGTTCTGA
Protein
MASSFLGNLVESTYLPLETATTILAMAGLFKWPPQATVNDGDRFDFIVIGSGVGAVIANRLTENEDVRVLLIEAGKNPSVESMLPGLFILLQNSYQDWNYVSEPEEATKNQQFGAYRTSAGKCLGGSSNINHFIHLRGDPCDFDSWAAYLKDESWSYKNVLPYFRKSETIQDEDILKYYANFHGVDGPVIITRQPDDSTRNIMESFEEIGVPSVLDLNTNNTVGFTESSFIIGNGRRQSTSQAYLNNLNRDNLYVLTETVAEKIIFEDNVAVGVILRLGSGEKITVYANREVIVSAGTFNPLTSSGF
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9IS42
C7EPE2
A0A0L7L247
A0A2A4IZ69
A0A2W1BQ09
A0A2A4JWF9
+ More
A0A2W1BRA5 A0A2H1WBV8 A0A2H1VCU6 A0A2H1WPH8 A0A2A4JE08 A0A2H1VZZ7 A0A2W1BEK9 A0A2H1VCV1 A0A2H1VP10 A0A212F6I8 A0A0N1PIV1 A0A194QAM1 A0A212FMS7 A0A194QC04 A0A3S2N607 A0A2H1VB55 A0A0N0PDR9 A0A3S2LRK7 A0A2H1W0I8 A0A2H1WQ34 A0A2W1BM44 A0A2W1BPR7 A0A2A4JYC1 A0A3S2LM78 D0ABA6 A0A2A4J139 A0A2A4JF29 A0A2W1BU23 A0A0L7LL99 A0A2H1W7M4 A0A194QAR0 A0A0L7KN60 A0A2H1W336 I4DQ20 A0A2H1VDD9 A0A2H1V4R2 A0A2H1VDA1 A0A212F6I0 A0A2W1BIV7 A0A2A4IZE5 A0A0L7LUK3 A0A0N1I9P3 A0A1B6JHF0 A0A1B6ISL6 A0A2H1V0S8 A0A2A4JSK9 A0A0N1IPT1 A0A194PPA0 A0A1B6HLV3 Q95NZ0 A0A2H1W3T3 A0A0N0PE61 A0A194R898 A0A2H1V9N6 H9J869 A0A2W1BWE7 A0A088S3T2 A0A194QGE4 H9JLP6 A0A194QNE6 A0A088S3S6 A0A088S3M8 A0A1B6HJ69 A0A088S5A0 A0A088SM07 A0A2A4JSJ8 A0A194PSS7 A0A291P0W5 A0A2W1BHP2 H9JTY7 A0A088SM03 A0A088S3M1 H6AGY0 A0A088S376 A0A194QAX1 V9IE09 A0A2A4JUI0 A0A2H1VJT7 A0A0C9RN66 A0A1Y9H2C4 A0A212FF54 K7J277 A0A2P8ZHS7 A0A1B6FPZ2 A0A0L7LPB4 A0A1B6KE48 A0A194PXZ3 A0A232FFY1 A0A1B6EWM9 A0A1B6EYD4 A0A023EVK5 A0A1B6MBY6 A0A2J7PXM4 A0A2A4K5W1
A0A2W1BRA5 A0A2H1WBV8 A0A2H1VCU6 A0A2H1WPH8 A0A2A4JE08 A0A2H1VZZ7 A0A2W1BEK9 A0A2H1VCV1 A0A2H1VP10 A0A212F6I8 A0A0N1PIV1 A0A194QAM1 A0A212FMS7 A0A194QC04 A0A3S2N607 A0A2H1VB55 A0A0N0PDR9 A0A3S2LRK7 A0A2H1W0I8 A0A2H1WQ34 A0A2W1BM44 A0A2W1BPR7 A0A2A4JYC1 A0A3S2LM78 D0ABA6 A0A2A4J139 A0A2A4JF29 A0A2W1BU23 A0A0L7LL99 A0A2H1W7M4 A0A194QAR0 A0A0L7KN60 A0A2H1W336 I4DQ20 A0A2H1VDD9 A0A2H1V4R2 A0A2H1VDA1 A0A212F6I0 A0A2W1BIV7 A0A2A4IZE5 A0A0L7LUK3 A0A0N1I9P3 A0A1B6JHF0 A0A1B6ISL6 A0A2H1V0S8 A0A2A4JSK9 A0A0N1IPT1 A0A194PPA0 A0A1B6HLV3 Q95NZ0 A0A2H1W3T3 A0A0N0PE61 A0A194R898 A0A2H1V9N6 H9J869 A0A2W1BWE7 A0A088S3T2 A0A194QGE4 H9JLP6 A0A194QNE6 A0A088S3S6 A0A088S3M8 A0A1B6HJ69 A0A088S5A0 A0A088SM07 A0A2A4JSJ8 A0A194PSS7 A0A291P0W5 A0A2W1BHP2 H9JTY7 A0A088SM03 A0A088S3M1 H6AGY0 A0A088S376 A0A194QAX1 V9IE09 A0A2A4JUI0 A0A2H1VJT7 A0A0C9RN66 A0A1Y9H2C4 A0A212FF54 K7J277 A0A2P8ZHS7 A0A1B6FPZ2 A0A0L7LPB4 A0A1B6KE48 A0A194PXZ3 A0A232FFY1 A0A1B6EWM9 A0A1B6EYD4 A0A023EVK5 A0A1B6MBY6 A0A2J7PXM4 A0A2A4K5W1
Pubmed
EMBL
BABH01024325
BABH01024320
GQ285072
ACT66690.2
JTDY01003559
KOB69344.1
+ More
NWSH01004322 PCG65227.1 KZ150085 PZC73783.1 NWSH01000445 PCG76355.1 KZ149986 PZC75677.1 ODYU01007560 SOQ50436.1 ODYU01001603 SOQ38054.1 ODYU01010073 SOQ54917.1 NWSH01001885 PCG69794.1 ODYU01005459 SOQ46343.1 KZ150336 PZC71326.1 ODYU01001607 SOQ38064.1 ODYU01003587 SOQ42538.1 AGBW02010007 OWR49347.1 KQ460044 KPJ18290.1 KQ459232 KPJ02522.1 AGBW02007651 OWR55045.1 KPJ02520.1 RSAL01000341 RVE42398.1 SOQ38065.1 KPJ18288.1 RVE42399.1 SOQ46342.1 ODYU01010216 SOQ55185.1 PZC73780.1 KZ149963 PZC76281.1 NWSH01000427 PCG76483.1 RSAL01003283 RVE40310.1 FP102341 CBH09301.1 NWSH01004424 PCG65132.1 NWSH01001625 PCG70657.1 PZC76280.1 JTDY01000758 KOB75991.1 ODYU01006844 SOQ49058.1 KPJ02519.1 JTDY01008771 KOB64409.1 ODYU01006017 SOQ47491.1 AK403892 BAM20010.1 ODYU01001944 SOQ38845.1 ODYU01000650 SOQ35789.1 ODYU01001935 SOQ38829.1 OWR49345.1 KZ150015 PZC74989.1 NWSH01004349 PCG65195.1 JTDY01000066 KOB79079.1 KPJ18287.1 GECU01009062 JAS98644.1 GECU01017785 JAS89921.1 ODYU01000155 SOQ34448.1 NWSH01000760 PCG74382.1 KPJ18289.1 KQ459603 KPI92960.1 GECU01032039 JAS75667.1 AY035784 AY035785 AAK56551.1 AAK56552.1 SOQ47492.1 KPJ18306.1 KQ460627 KPJ13480.1 ODYU01001405 SOQ37539.1 BABH01039860 BABH01039861 KZ149916 PZC77944.1 KF717661 AIO02863.1 KPJ02521.1 BABH01005285 KQ461194 KPJ06879.1 KF717651 AIO02853.1 KF717664 AIO02866.1 GECU01033045 JAS74661.1 KF717655 AIO02857.1 KF717656 KF717657 KF717660 KF717667 AIO02858.1 AIO02859.1 AIO02862.1 AIO02869.1 PCG74380.1 KQ459595 KPI96023.1 MF687606 ATJ44532.1 PZC73781.1 BABH01003798 BABH01003799 KF717652 AIO02854.1 KF717654 AIO02856.1 JF433972 KF717653 KF717659 KF717662 KF717663 KF717665 KF717666 KF717668 AEM17059.1 AIO02855.1 AIO02861.1 AIO02864.1 AIO02865.1 AIO02867.1 AIO02868.1 AIO02870.1 KF717658 AIO02860.1 KPJ02688.1 JR040861 AEY59320.1 NWSH01000559 PCG75661.1 ODYU01002782 SOQ40702.1 GBYB01008526 JAG78293.1 AGBW02008849 OWR52369.1 AAZX01008181 PYGN01000053 PSN56054.1 GECZ01017548 JAS52221.1 JTDY01000418 KOB77282.1 GEBQ01030254 JAT09723.1 KQ459585 KPI98211.1 NNAY01000267 OXU29612.1 GECZ01027417 JAS42352.1 GECZ01026787 JAS42982.1 GAPW01000526 JAC13072.1 GEBQ01006583 JAT33394.1 NEVH01020853 PNF21087.1 NWSH01000095 PCG79621.1
NWSH01004322 PCG65227.1 KZ150085 PZC73783.1 NWSH01000445 PCG76355.1 KZ149986 PZC75677.1 ODYU01007560 SOQ50436.1 ODYU01001603 SOQ38054.1 ODYU01010073 SOQ54917.1 NWSH01001885 PCG69794.1 ODYU01005459 SOQ46343.1 KZ150336 PZC71326.1 ODYU01001607 SOQ38064.1 ODYU01003587 SOQ42538.1 AGBW02010007 OWR49347.1 KQ460044 KPJ18290.1 KQ459232 KPJ02522.1 AGBW02007651 OWR55045.1 KPJ02520.1 RSAL01000341 RVE42398.1 SOQ38065.1 KPJ18288.1 RVE42399.1 SOQ46342.1 ODYU01010216 SOQ55185.1 PZC73780.1 KZ149963 PZC76281.1 NWSH01000427 PCG76483.1 RSAL01003283 RVE40310.1 FP102341 CBH09301.1 NWSH01004424 PCG65132.1 NWSH01001625 PCG70657.1 PZC76280.1 JTDY01000758 KOB75991.1 ODYU01006844 SOQ49058.1 KPJ02519.1 JTDY01008771 KOB64409.1 ODYU01006017 SOQ47491.1 AK403892 BAM20010.1 ODYU01001944 SOQ38845.1 ODYU01000650 SOQ35789.1 ODYU01001935 SOQ38829.1 OWR49345.1 KZ150015 PZC74989.1 NWSH01004349 PCG65195.1 JTDY01000066 KOB79079.1 KPJ18287.1 GECU01009062 JAS98644.1 GECU01017785 JAS89921.1 ODYU01000155 SOQ34448.1 NWSH01000760 PCG74382.1 KPJ18289.1 KQ459603 KPI92960.1 GECU01032039 JAS75667.1 AY035784 AY035785 AAK56551.1 AAK56552.1 SOQ47492.1 KPJ18306.1 KQ460627 KPJ13480.1 ODYU01001405 SOQ37539.1 BABH01039860 BABH01039861 KZ149916 PZC77944.1 KF717661 AIO02863.1 KPJ02521.1 BABH01005285 KQ461194 KPJ06879.1 KF717651 AIO02853.1 KF717664 AIO02866.1 GECU01033045 JAS74661.1 KF717655 AIO02857.1 KF717656 KF717657 KF717660 KF717667 AIO02858.1 AIO02859.1 AIO02862.1 AIO02869.1 PCG74380.1 KQ459595 KPI96023.1 MF687606 ATJ44532.1 PZC73781.1 BABH01003798 BABH01003799 KF717652 AIO02854.1 KF717654 AIO02856.1 JF433972 KF717653 KF717659 KF717662 KF717663 KF717665 KF717666 KF717668 AEM17059.1 AIO02855.1 AIO02861.1 AIO02864.1 AIO02865.1 AIO02867.1 AIO02868.1 AIO02870.1 KF717658 AIO02860.1 KPJ02688.1 JR040861 AEY59320.1 NWSH01000559 PCG75661.1 ODYU01002782 SOQ40702.1 GBYB01008526 JAG78293.1 AGBW02008849 OWR52369.1 AAZX01008181 PYGN01000053 PSN56054.1 GECZ01017548 JAS52221.1 JTDY01000418 KOB77282.1 GEBQ01030254 JAT09723.1 KQ459585 KPI98211.1 NNAY01000267 OXU29612.1 GECZ01027417 JAS42352.1 GECZ01026787 JAS42982.1 GAPW01000526 JAC13072.1 GEBQ01006583 JAT33394.1 NEVH01020853 PNF21087.1 NWSH01000095 PCG79621.1
Proteomes
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9IS42
C7EPE2
A0A0L7L247
A0A2A4IZ69
A0A2W1BQ09
A0A2A4JWF9
+ More
A0A2W1BRA5 A0A2H1WBV8 A0A2H1VCU6 A0A2H1WPH8 A0A2A4JE08 A0A2H1VZZ7 A0A2W1BEK9 A0A2H1VCV1 A0A2H1VP10 A0A212F6I8 A0A0N1PIV1 A0A194QAM1 A0A212FMS7 A0A194QC04 A0A3S2N607 A0A2H1VB55 A0A0N0PDR9 A0A3S2LRK7 A0A2H1W0I8 A0A2H1WQ34 A0A2W1BM44 A0A2W1BPR7 A0A2A4JYC1 A0A3S2LM78 D0ABA6 A0A2A4J139 A0A2A4JF29 A0A2W1BU23 A0A0L7LL99 A0A2H1W7M4 A0A194QAR0 A0A0L7KN60 A0A2H1W336 I4DQ20 A0A2H1VDD9 A0A2H1V4R2 A0A2H1VDA1 A0A212F6I0 A0A2W1BIV7 A0A2A4IZE5 A0A0L7LUK3 A0A0N1I9P3 A0A1B6JHF0 A0A1B6ISL6 A0A2H1V0S8 A0A2A4JSK9 A0A0N1IPT1 A0A194PPA0 A0A1B6HLV3 Q95NZ0 A0A2H1W3T3 A0A0N0PE61 A0A194R898 A0A2H1V9N6 H9J869 A0A2W1BWE7 A0A088S3T2 A0A194QGE4 H9JLP6 A0A194QNE6 A0A088S3S6 A0A088S3M8 A0A1B6HJ69 A0A088S5A0 A0A088SM07 A0A2A4JSJ8 A0A194PSS7 A0A291P0W5 A0A2W1BHP2 H9JTY7 A0A088SM03 A0A088S3M1 H6AGY0 A0A088S376 A0A194QAX1 V9IE09 A0A2A4JUI0 A0A2H1VJT7 A0A0C9RN66 A0A1Y9H2C4 A0A212FF54 K7J277 A0A2P8ZHS7 A0A1B6FPZ2 A0A0L7LPB4 A0A1B6KE48 A0A194PXZ3 A0A232FFY1 A0A1B6EWM9 A0A1B6EYD4 A0A023EVK5 A0A1B6MBY6 A0A2J7PXM4 A0A2A4K5W1
A0A2W1BRA5 A0A2H1WBV8 A0A2H1VCU6 A0A2H1WPH8 A0A2A4JE08 A0A2H1VZZ7 A0A2W1BEK9 A0A2H1VCV1 A0A2H1VP10 A0A212F6I8 A0A0N1PIV1 A0A194QAM1 A0A212FMS7 A0A194QC04 A0A3S2N607 A0A2H1VB55 A0A0N0PDR9 A0A3S2LRK7 A0A2H1W0I8 A0A2H1WQ34 A0A2W1BM44 A0A2W1BPR7 A0A2A4JYC1 A0A3S2LM78 D0ABA6 A0A2A4J139 A0A2A4JF29 A0A2W1BU23 A0A0L7LL99 A0A2H1W7M4 A0A194QAR0 A0A0L7KN60 A0A2H1W336 I4DQ20 A0A2H1VDD9 A0A2H1V4R2 A0A2H1VDA1 A0A212F6I0 A0A2W1BIV7 A0A2A4IZE5 A0A0L7LUK3 A0A0N1I9P3 A0A1B6JHF0 A0A1B6ISL6 A0A2H1V0S8 A0A2A4JSK9 A0A0N1IPT1 A0A194PPA0 A0A1B6HLV3 Q95NZ0 A0A2H1W3T3 A0A0N0PE61 A0A194R898 A0A2H1V9N6 H9J869 A0A2W1BWE7 A0A088S3T2 A0A194QGE4 H9JLP6 A0A194QNE6 A0A088S3S6 A0A088S3M8 A0A1B6HJ69 A0A088S5A0 A0A088SM07 A0A2A4JSJ8 A0A194PSS7 A0A291P0W5 A0A2W1BHP2 H9JTY7 A0A088SM03 A0A088S3M1 H6AGY0 A0A088S376 A0A194QAX1 V9IE09 A0A2A4JUI0 A0A2H1VJT7 A0A0C9RN66 A0A1Y9H2C4 A0A212FF54 K7J277 A0A2P8ZHS7 A0A1B6FPZ2 A0A0L7LPB4 A0A1B6KE48 A0A194PXZ3 A0A232FFY1 A0A1B6EWM9 A0A1B6EYD4 A0A023EVK5 A0A1B6MBY6 A0A2J7PXM4 A0A2A4K5W1
PDB
3T37
E-value=1.51746e-25,
Score=287
Ontologies
GO
Topology
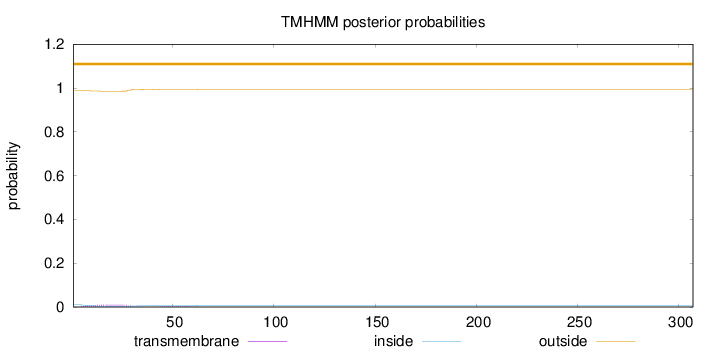
Length:
307
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.285640000000001
Exp number, first 60 AAs:
0.27996
Total prob of N-in:
0.01274
outside
1 - 307
Population Genetic Test Statistics
Pi
189.176535
Theta
161.283872
Tajima's D
0.953079
CLR
0
CSRT
0.650567471626419
Interpretation
Uncertain
