Gene
KWMTBOMO14489
Annotation
reverse_transcriptase_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.175 Nuclear Reliability : 1.508
Sequence
CDS
ATGGCTGGTTTCCGAGCGCTATCTCATCAACCTACGAGCTCTGATGGTGAAGTTAAAAAAAAAATTAGAGGGTACCTCCCGCTCCGCGGGGGAGAATCCCTCCGTGCGGTCGAGCGATCGGCCGCACGCTGCCCCACCGTTTCTGGGGGGGGCAAACATTGCCACGACGAAGGGGGCGACAGCACGGGTCGCGTAAGGGACAGAGTCCCCACCGTGCGCCTCGAACGCTGCGAGAGCCTGATCTCGCTCTACTCGGCGCGGAGCTCCGCAAGGGGCGACGCGTCCAGTGGGCGGGAGACGCCAGCGCTGTTCGTGGACAGTGCTCGACAGCGCACTAGACGGAAGAGGAAGTCGGCTCGTCAGCCGGCATCTGTCCCTCCGTCCGACTCCTCGGAAGGAGAGCCTACCAGCGCCAAGTACTTGGCGTTGGCGGAAGGGCCGGCGACTGCTGCGGCGTCGCCCGCCACCGGCTTTGACAAAAGAGCATCTTGCGGACTGGATGCTGCCTCCTCGGCGCAGTCCAGGGCCGCCTGCCGTGAGGCACATAAACGGTTGGCAACGGTTGGAGAGGTCGAGCGACTGGAGCGCTCCCTGACAGAGGTGGGTCCCTATGACCCCCTCCGTAGATATGGCATCAGCCAAAAAATTGTTTCTACGGTGGCGACGAAGTCGAGTGCAGGCCTAAAGGGCGACTATGTGCGGGCCCTAAAATCGGCCGCCTCCTCCATGGAGGAAGTAATGGTGGAGGTGTTGGAGCGCACCAAGGATGGGGAGACGGAAGGTCTGCGCAAATCGGTGGACCTGCTGCACGCCCGACTGGCCCAAATCAGCGCGGAGAACGGTCATCTTCGCGCTGAGAACGGGCAGATGAAGGTGGACTTGGCCGAGTTGCGCACACTGGTCAACGACCTAATCGAGAAGCAGTGCAACTCGGCCCCTGTACCTACTCCGCTCGAGACCGAAGAGGCCAATGAGGCCGAGGAGCTGAGGCGGCAGCTCCTAATACAGGAGGCACGTAGCTCCAACGTGGAGCGCGCGAGACCGCCCCTCGCCCATGAAGCGAAAAGCAGTGCGCCTACCTACACCTGCGGCTGCGGGGGTGCCGGCACCCACGGCCCCTACAAGAAAACCGGCGTCCGCTCCTGCCCCGCCCCAGCCTGCGAAAGCAGGGCCCGGCAGGAGCCGGACAAAGAAGAAGGGAGGCGTCAACGCCGCCAAACCGGCACTGACCCCCCAGCCCGCCCACTGCCCCCCGCCCCGGCCAACATGGATGAGGCCTGGACGACGGTAGTGCGGCGTGGGCCCAAAAAGGTGGCGGCGCCTCCTGCGCCACGCCCCAAGCCCACCCCTGCGGCCGCCGCTCCCCGCCAAGAGGGTCGAGCGGAGCCGAGGGGCCGAACTGGTAGAAGGAGGCCGCGCGCCCCGCGGTCGGCTGCAGTGGTAGTGGAGCTGCTGCCGGCCGGCAAAGAAAAGGGCATCACCTACCACGAGGTGATGACCCAGGCGCGTGGCGGCGTGAATGTGGACGCCCTCGGCGTGGAGGGGGGCATCAAGGTCCGGCAGACGGCCAACGGGGCTCGTCTTATAGAGTGCCCCGGCCCCAACACAAACGCCGCGGCCGAAAAATTGGCGGCCCGGCTGAGGGAGATCCTGCCGGACCCCGAGGTGGTGCGCATCGCGCGGCCCGTGAAGATGGGGGAAGTAAAAATCACGGGCCTCGACGAGTGCGCCACGAAAGAGGAGGTCGCCGCGGCCATCGCGTCGCAGGGCAACTGCGCCCTCGCCGACGTCAAAGTCGGGGAGCTCCGGGCGACGTACTCCGGAACCCGGACGGCGTGGGCGCGTTGCCCGGTAGTAACGGCGAACCTCCTGGCCACCCCTCCGCAGGGTCGGCCTTCGGACAGCCCTGGATGGCTGCGCGTGGGCTGGGTAGTGGCCCACGTGCAACTGCAGGAGAGCAGGCCGTGGCGTTGTCTTCGGTGCTTCGGCACCGGCCACGGCCTCGCCAAGTGCCCCTCGACCGTGGACCGCAGCGATCTGTGTTTCCGCTGCGGCCAACCGGGGCACAAGGCAGCCTCCTGCACCACAGCCGCGCCGCACTGCGTCTTGTGCGACGCGGCTAAAAGGAAGGCCGACCACCGGGCCGGGGGCCCGGCGTGTAAGTCCGCCCCCTCCTCTACCAAAACGAGGAGGGGCGGCAAGAAAAATAAAAAGAAGGAGAGGCCGGCTGCAATAAGGGCAGCCGGCGAGTCAGTTCCCGCCGCGGCACCAGACGAGCCGCAGGGCGGGACAGACGAGGGAGCGATGGACGTAGCCCCGAGCCTCGCCGAGTTCGAGCGGTTTCTGGGCGGGCTCGAGGCGCTTTTACACCGCCTCGGGTCCCGCCCTGTCATCGTCGCGGGGGACTTCAATGCCAAGTGCACGGCTTGGGGTTCCCCCCGCACGGATGCGCGAGGCGAGGCCCTTTCGGAGTGGGCGATCGCGACCGGCCTCTGTCTCCTAAACAGAGGCACGGTCGCGACTTGCGTGCGGTGGAACGGGGAATCTCACGTGGACGTGACGTTCGCGTCCCCGTCCGCCGTGCGCCGCACCCGCGGCTGGCGCGTACTTGAGGGGGCGGAGACGCTCTCGGACCACAGATACGTCCGATTCGAGCTCTCCGCCTCCTCCTTGTCGTCGTCGTCAGCCGCGGGCGCCCGCGGTGAGGAGGAGCTCCCGCAAGGTGCGCCCCGTTCGTTCCCCAGGTGGGCCTTGAAACGAATGAACAAGGAGTTGCTGATGGAGGCGGCCGCTGTTGCTGCGTGGGCGCCAAAACCCGCGCGGCTCGCGGACGTGGATGCGGAGGTCGACTGTTTCCGGGGCACCATGGCAAACATTTGTGATGCCGCCATGCCCCGGGTCGGCCCCGAGCACCACGTGGGGGTGCGTTCTGGTGGTCGCCCGAGATCGCGAGACTCCACGGCGGCCCGGCTGCACGCTGACTGTCGTCAAAAGCAGACGGCGCTGCAGCTGGCCATCGGCGAGGCCAAGAAGCAGAGCATGAAGACTCTCCTGGAGTCGCTCGACGAAGATCCCTGGGGGCGCCCATACAAGATGGTTCGCAGGAAACTGCAGCCGTGGGCGCTCCCGGTGACCGAGCGGCTCCAGCCTCGGCAGCTGCGGGAAATTGTTACGGCACTCTTCCCGCCGGCAGCGGGGGGGGACTTTGAGCCCCCCCAGATGGACGCCACGTGGGGCACGCCTCCGCGTCGCCCCAGCGTTCCCGCTGCCGAGGAGCCCCCTCCCCCTATCACGGGGGCGGAGATCCATGCGGCCGTGTCCAGAATGAGAGCGAAGGACGCGGCCCCCGGCCCTGACGGCGTTCACGGCCGGGTCTGGAGCTTGGCCTTCGAGGCCCTAGGGGACCGGCTCGGGGGGCTTTTCGAAGCGTGCCTCGAGTCGGGACGGTTCCCGTCGAAATGGAAGACGGGCAGACTGGTCCTTCTGCGGAAGGACGGGCGCCCCGCGGACTCACCCGCGGGCTATCGCCCCATCGTGTTGCTGGACGAGGCGGGCAAGATGCTCGAGAGGATCGTCGCGGCCCGCATCGTCCGGCACCTGACCGAGACGGGTCCCGATCTTTCGGCGGAGCAATACGGCTTCAGAGAGGGCCGCTCGACTATCGACGCAATTCTGCGCGTGCGGGCCCTCTCCGACGAAGCCGTATCCCGGGGCGGGGTGGCGATGGCGTTATCCTTAGATGTAAGGAACGCCTTCAACACCCTGCCCTGGTCGGTGATCGCGGGGGCGCTGGAGTATCACGGCGTCCCCGCATACCTCCGCCGACTGATCGGCTCCTATCTCGAGGGCAGGTCGATCCAGTGCATCGGACACGGTGGAGTGATGTACCGCTTCCCCGTCGAGCGCGGTGTTCCGCAGGGGTCCGTCCTGGGCCCCCTGCTGTGGAACATCGGCTACGACTGGGTCCTGCGGGGCGTCATTCGGGGACCCCTCCCCGGGCTCAGCGTAATTTGTTACGCTGACGACACGTTGGTCGTAGCTCCGGGGAGGGACTACCGGGAGTCTGCCCGTCTGGCGTGCGCAGGCGTGGCACACGTCGTCACCAGGATCCGACGGTTGGGGCTCGAGGTGGCGCTCGACAAAACCCAGGCCCTGATGTTTCACGGGCCGGGACGAGCGCCGCCTGTGGGTGCCCACCTCGTGATCGGAGGCGTCCGCGTCGGGGTCGGGGTGACCGGTCTTCGGTACCTCGGCCTTGAACTGGATGGTCGGTGGAACTTCCGCGCTCACTTTGCGAAGTTGGGCCCTCGACTGATGGCGACGGCCGGCTCTTTGAGCCGGCTGCTTCCAAACGTCGGGGGTCCCGATCAGGTGGCGCGCCGCCTCTACATGGGGGTGGTGCGGTCGATGGCACTATACGGCGCGCCCGTATGGTGCCACGCCCTGACCCGCCAGAACGTCGCCGCGCTGCGACGTCCGCAGCGCGCGATCGCGGTCAGGGCGATCCGAGGATACCGCACCGTCTCCTTCGAGGCGGCGTGCTTGCTCGCCGGGGCGCCACCCTGGGACCTGGAGGCGGAGGCGCTCGCTGCCGATTACCGGTGGCGTAGCGACCTCCGCTCTAGGGGGAAGGGCGCCCCAGCGAAGATAGCCGGAAGGGAACCGACGGCGCAGTGCCACCACTGCGCGGACCGCGACGAGGAAGACACAGCGGAACACACGCTGGCGCGTTGCTCTGGATTCGACGAGCAACGCGCCGCCCTCGTCGCGGTCATTGGAGAGGACCTCTCGCTGCCGCGCGTCGTGGCTACGATGCTCGGCAGCGACGCGTCCTGGAAGGCGATGCTCGACTTCTGCGAGTCCACCATCTCGTAG
Protein
MAGFRALSHQPTSSDGEVKKKIRGYLPLRGGESLRAVERSAARCPTVSGGGKHCHDEGGDSTGRVRDRVPTVRLERCESLISLYSARSSARGDASSGRETPALFVDSARQRTRRKRKSARQPASVPPSDSSEGEPTSAKYLALAEGPATAAASPATGFDKRASCGLDAASSAQSRAACREAHKRLATVGEVERLERSLTEVGPYDPLRRYGISQKIVSTVATKSSAGLKGDYVRALKSAASSMEEVMVEVLERTKDGETEGLRKSVDLLHARLAQISAENGHLRAENGQMKVDLAELRTLVNDLIEKQCNSAPVPTPLETEEANEAEELRRQLLIQEARSSNVERARPPLAHEAKSSAPTYTCGCGGAGTHGPYKKTGVRSCPAPACESRARQEPDKEEGRRQRRQTGTDPPARPLPPAPANMDEAWTTVVRRGPKKVAAPPAPRPKPTPAAAAPRQEGRAEPRGRTGRRRPRAPRSAAVVVELLPAGKEKGITYHEVMTQARGGVNVDALGVEGGIKVRQTANGARLIECPGPNTNAAAEKLAARLREILPDPEVVRIARPVKMGEVKITGLDECATKEEVAAAIASQGNCALADVKVGELRATYSGTRTAWARCPVVTANLLATPPQGRPSDSPGWLRVGWVVAHVQLQESRPWRCLRCFGTGHGLAKCPSTVDRSDLCFRCGQPGHKAASCTTAAPHCVLCDAAKRKADHRAGGPACKSAPSSTKTRRGGKKNKKKERPAAIRAAGESVPAAAPDEPQGGTDEGAMDVAPSLAEFERFLGGLEALLHRLGSRPVIVAGDFNAKCTAWGSPRTDARGEALSEWAIATGLCLLNRGTVATCVRWNGESHVDVTFASPSAVRRTRGWRVLEGAETLSDHRYVRFELSASSLSSSSAAGARGEEELPQGAPRSFPRWALKRMNKELLMEAAAVAAWAPKPARLADVDAEVDCFRGTMANICDAAMPRVGPEHHVGVRSGGRPRSRDSTAARLHADCRQKQTALQLAIGEAKKQSMKTLLESLDEDPWGRPYKMVRRKLQPWALPVTERLQPRQLREIVTALFPPAAGGDFEPPQMDATWGTPPRRPSVPAAEEPPPPITGAEIHAAVSRMRAKDAAPGPDGVHGRVWSLAFEALGDRLGGLFEACLESGRFPSKWKTGRLVLLRKDGRPADSPAGYRPIVLLDEAGKMLERIVAARIVRHLTETGPDLSAEQYGFREGRSTIDAILRVRALSDEAVSRGGVAMALSLDVRNAFNTLPWSVIAGALEYHGVPAYLRRLIGSYLEGRSIQCIGHGGVMYRFPVERGVPQGSVLGPLLWNIGYDWVLRGVIRGPLPGLSVICYADDTLVVAPGRDYRESARLACAGVAHVVTRIRRLGLEVALDKTQALMFHGPGRAPPVGAHLVIGGVRVGVGVTGLRYLGLELDGRWNFRAHFAKLGPRLMATAGSLSRLLPNVGGPDQVARRLYMGVVRSMALYGAPVWCHALTRQNVAALRRPQRAIAVRAIRGYRTVSFEAACLLAGAPPWDLEAEALAADYRWRSDLRSRGKGAPAKIAGREPTAQCHHCADRDEEDTAEHTLARCSGFDEQRAALVAVIGEDLSLPRVVATMLGSDASWKAMLDFCESTIS
Summary
Proteomes
Interpro
Gene 3D
PDB
1WDU
E-value=3.69033e-08,
Score=144
Ontologies
GO
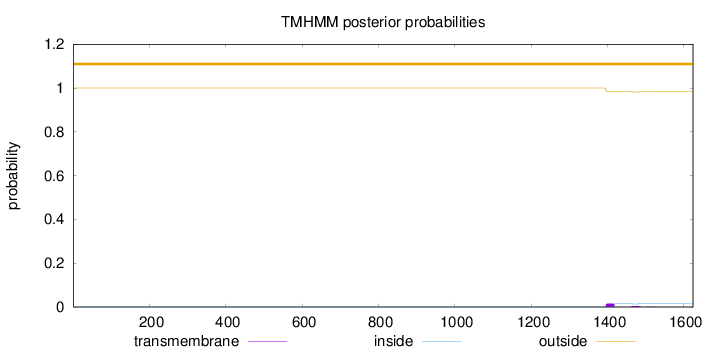
Topology
Length:
1624
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.45018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00000
outside
1 - 1624
Population Genetic Test Statistics
Pi
14.460474
Theta
148.741946
Tajima's D
-1.512945
CLR
1439.767789
CSRT
0.0585970701464927
Interpretation
Uncertain
