Pre Gene Modal
BGIBMGA000155
Annotation
PREDICTED:_methylosome_subunit_pICln_[Amyelois_transitella]
Full name
Methylosome subunit pICln
Alternative Name
Chloride channel, nucleotide sensitive 1A
Chloride conductance regulatory protein ICln
Chloride ion current inducer protein
Reticulocyte pICln
Chloride conductance regulatory protein ICln
Chloride ion current inducer protein
Reticulocyte pICln
Location in the cell
Nuclear Reliability : 2.696
Sequence
CDS
ATGGTAGTTGTGTCAAGTAATTTTGCTGAGCCGGCCGATGGTGTGTTGCTACAGTCTCCATCTACAAAACTTCTCATAAATGATCAAGAATTAGGCACAGCTACGTTGTACATCACTGAAAACAACGTCATATGGGGCGGAGGCGTTAGTCCATCAGGCAATCCAGCTCCGACCATCAATCTACTCTATCCAAGCATATCTCTACATGCGATACAACGCGAACCATCCCCTGCCTTGTACATGGTTCTTAATTATGAGTTACGTTTACCAGAACTCAGTCAGCAAGCAGGTTCCACTGATGCCGACGACGATGATGACTTTGATGTCGATGATCAACCCATAACACAGCTCAGATTCATCCCAGAAAACGAGAACGAATTACAAGCCATGTATTCGGCCATGTGTCAGGGACAAGAATTGCATCCGGATCCGGCTGATGAGGTCGAGGATGATGACCCATACATGGACGGAGAGGAGTTTGATGAAGGGGAGGAGGAGTTTGAAGATGCTGAGGAGAGCGCGCCCGACCCCGCCGCCGCGCTGAGACGCATGAGGCTCGGCCCCCCGCCCGGTGGGTCCTCGTTATAG
Protein
MVVVSSNFAEPADGVLLQSPSTKLLINDQELGTATLYITENNVIWGGGVSPSGNPAPTINLLYPSISLHAIQREPSPALYMVLNYELRLPELSQQAGSTDADDDDDFDVDDQPITQLRFIPENENELQAMYSAMCQGQELHPDPADEVEDDDPYMDGEEFDEGEEEFEDAEESAPDPAAALRRMRLGPPPGGSSL
Summary
Description
Involved in both the assembly of spliceosomal snRNPs and the methylation of Sm proteins (PubMed:21081503, PubMed:18984161). Chaperone that regulates the assembly of spliceosomal U1, U2, U4 and U5 small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome. Thereby, plays an important role in the splicing of cellular pre-mRNAs. Most spliceosomal snRNPs contain a common set of Sm proteins SNRPB, SNRPD1, SNRPD2, SNRPD3, SNRPE, SNRPF and SNRPG that assemble in a heptameric protein ring on the Sm site of the small nuclear RNA to form the core snRNP. In the cytosol, the Sm proteins SNRPD1, SNRPD2, SNRPE, SNRPF and SNRPG are trapped in an inactive 6S pICln-Sm complex by the chaperone CLNS1A that controls the assembly of the core snRNP. Dissociation by the SMN complex of CLNS1A from the trapped Sm proteins and their transfer to an SMN-Sm complex triggers the assembly of core snRNPs and their transport to the nucleus. May also indirectly participate in cellular volume control by activation of a swelling-induced chloride conductance pathway.
Involved in both the assembly of spliceosomal snRNPs and the methylation of Sm proteins. Chaperone that regulates the assembly of spliceosomal U1, U2, U4 and U5 small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome (By similarity). Thereby, plays an important role in the splicing of cellular pre-mRNAs. Most spliceosomal snRNPs contain a common set of Sm proteins SNRPB, SNRPD1, SNRPD2, SNRPD3, SNRPE, SNRPF and SNRPG that assemble in a heptameric protein ring on the Sm site of the small nuclear RNA to form the core snRNP. In the cytosol, the Sm proteins SNRPD1, SNRPD2, SNRPE, SNRPF and SNRPG are trapped in an inactive 6S pICln-Sm complex by the chaperone CLNS1A that controls the assembly of the core snRNP. Dissociation by the SMN complex of CLNS1A from the trapped Sm proteins and their transfer to an SMN-Sm complex triggers the assembly of core snRNPs and their transport to the nucleus. May also indirectly participate in cellular volume control by activation of a swelling-induced chloride conductance pathway.
Involved in both the assembly of spliceosomal snRNPs and the methylation of Sm proteins. Chaperone that regulates the assembly of spliceosomal U1, U2, U4 and U5 small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome (By similarity). Thereby, plays an important role in the splicing of cellular pre-mRNAs. Most spliceosomal snRNPs contain a common set of Sm proteins SNRPB, SNRPD1, SNRPD2, SNRPD3, SNRPE, SNRPF and SNRPG that assemble in a heptameric protein ring on the Sm site of the small nuclear RNA to form the core snRNP. In the cytosol, the Sm proteins SNRPD1, SNRPD2, SNRPE, SNRPF and SNRPG are trapped in an inactive 6S pICln-Sm complex by the chaperone CLNS1A that controls the assembly of the core snRNP. Dissociation by the SMN complex of CLNS1A from the trapped Sm proteins and their transfer to an SMN-Sm complex triggers the assembly of core snRNPs and their transport to the nucleus. May also indirectly participate in cellular volume control by activation of a swelling-induced chloride conductance pathway.
Subunit
Component of the methylosome, a 20S complex containing at least PRMT5/SKB1, WDR77/MEP50 and CLNS1A/pICln (PubMed:21081503, PubMed:11747828, PubMed:18984161). May mediate SNRPD1 and SNRPD3 methylation. Forms a 6S pICln-Sm complex composed of CLNS1A/pICln, SNRPD1, SNRPD2, SNRPE, SNRPF and SNRPG; ring-like structure where CLNS1A/pICln mimics additional Sm proteins and which is unable to assemble into the core snRNP (PubMed:18984161). Interacts with LSM10 and LSM11 (PubMed:16087681).
Component of the methylosome, a 20S complex containing at least PRMT5/SKB1, WDR77/MEP50 and CLNS1A/pICln. May mediate SNRPD1 and SNRPD3 methylation. Forms a 6S pICln-Sm complex composed of CLNS1A/pICln, SNRPD1, SNRPD2, SNRPE, SNRPF and SNRPG; ring-like structure where CLNS1A/pICln mimics additional Sm proteins and which is unable to assemble into the core snRNP. Interacts with LSM10 and LSM11 (By similarity).
Component of the methylosome, a 20S complex containing at least PRMT5/SKB1, WDR77/MEP50 and CLNS1A/pICln. May mediate SNRPD1 and SNRPD3 methylation. Forms a 6S pICln-Sm complex composed of CLNS1A/pICln, SNRPD1, SNRPD2, SNRPE, SNRPF and SNRPG; ring-like structure where CLNS1A/pICln mimics additional Sm proteins and which is unable to assemble into the core snRNP. Interacts with LSM10 and LSM11 (By similarity).
Similarity
Belongs to the pICln (TC 1.A.47) family.
Keywords
Acetylation
Complete proteome
Cytoplasm
Cytoskeleton
Direct protein sequencing
Nucleus
Phosphoprotein
Polymorphism
Reference proteome
Feature
chain Methylosome subunit pICln
Uniprot
H9ISC8
A0A212FFT5
A0A3S2M953
S4P810
A0A0N1ID77
A0A2H1WLW8
+ More
A0A2A4JJC2 A0A1E1VYK6 A0A1E1W1Y9 A0A087ZX28 A0A2A3EC88 A0A154PFB1 A0A067R0J2 A0A0M9A1L2 A0A310SJX7 E2AYH0 A0A2P8XTY0 A0A0J7KZR6 A0A2K5R0P8 A0A2R8MJB4 A0A026WP68 A0A2K5R0P5 U3FEP2 A0A2K5BXI8 A0A2K6FA31 A0A2K6FA49 A0A2K5BXI5 H0X015 A0A2K6CNB8 A0A1D5QSS3 A0A2K6LHR9 A0A2K5VED0 G3QKQ8 A0A2I3HPW9 A0A2K5ZYD1 A0A2R9B7Q6 A0A2I3MUM9 A0A2K6RQ17 A0A2J8V0T9 A0A2I3RSE1 E9PJF4 A0A218ULT2 A0A0P5X2I9 A0A0P5USH3 A0A0P4VLQ5 T1HI80 A0A2K5KD94 E9ISZ4 E2BUH9 A0A226N4G7 A0A0P5U5B7 A0A0P5A0D7 A0A0P5YVA2 G1S432 A0A2K6RPY5 A0A2K6CN69 A0A165A9N0 A0A2K6LHN5 A0A2I2Z6E0 A0A2K5ZY78 A0A0D9QUZ6 A0A2R9B7R3 A0A096N4Y3 A0A2J8V0T8 G8F520 G7NEE5 K7DM64 P54105 Q5R719 A0A2K5KD78 A0A0P4X774 A0A0P6JL91 A0A0P5DMC0 A0A2K5C8U2 K7IZ42 F4X3S6 A0A151XJB0 A0A3M0K2M9 W5MHL2 A0A3Q0EBK3 A0A195E023 A0A195F210 E9H646 A0A2K6L244 A0A1W4YJE6 A0A195BFI1 A0A3Q0E7E2 A0A158P0L7 A0A2J7QBP2 A0A2J7QBP5
A0A2A4JJC2 A0A1E1VYK6 A0A1E1W1Y9 A0A087ZX28 A0A2A3EC88 A0A154PFB1 A0A067R0J2 A0A0M9A1L2 A0A310SJX7 E2AYH0 A0A2P8XTY0 A0A0J7KZR6 A0A2K5R0P8 A0A2R8MJB4 A0A026WP68 A0A2K5R0P5 U3FEP2 A0A2K5BXI8 A0A2K6FA31 A0A2K6FA49 A0A2K5BXI5 H0X015 A0A2K6CNB8 A0A1D5QSS3 A0A2K6LHR9 A0A2K5VED0 G3QKQ8 A0A2I3HPW9 A0A2K5ZYD1 A0A2R9B7Q6 A0A2I3MUM9 A0A2K6RQ17 A0A2J8V0T9 A0A2I3RSE1 E9PJF4 A0A218ULT2 A0A0P5X2I9 A0A0P5USH3 A0A0P4VLQ5 T1HI80 A0A2K5KD94 E9ISZ4 E2BUH9 A0A226N4G7 A0A0P5U5B7 A0A0P5A0D7 A0A0P5YVA2 G1S432 A0A2K6RPY5 A0A2K6CN69 A0A165A9N0 A0A2K6LHN5 A0A2I2Z6E0 A0A2K5ZY78 A0A0D9QUZ6 A0A2R9B7R3 A0A096N4Y3 A0A2J8V0T8 G8F520 G7NEE5 K7DM64 P54105 Q5R719 A0A2K5KD78 A0A0P4X774 A0A0P6JL91 A0A0P5DMC0 A0A2K5C8U2 K7IZ42 F4X3S6 A0A151XJB0 A0A3M0K2M9 W5MHL2 A0A3Q0EBK3 A0A195E023 A0A195F210 E9H646 A0A2K6L244 A0A1W4YJE6 A0A195BFI1 A0A3Q0E7E2 A0A158P0L7 A0A2J7QBP2 A0A2J7QBP5
Pubmed
19121390
22118469
23622113
26354079
24845553
20798317
+ More
29403074 24508170 30249741 25243066 17431167 22398555 22722832 25362486 16136131 17081983 16554811 17487921 18691976 18669648 18318008 19413330 19369195 19690332 20068231 21269460 21406692 22814378 23186163 24275569 27129103 21282665 22002653 25319552 8579598 7887970 9359436 10825435 14702039 15489334 10330151 11747828 11713266 16087681 17525332 18984161 21081503 20075255 21719571 21292972 21347285
29403074 24508170 30249741 25243066 17431167 22398555 22722832 25362486 16136131 17081983 16554811 17487921 18691976 18669648 18318008 19413330 19369195 19690332 20068231 21269460 21406692 22814378 23186163 24275569 27129103 21282665 22002653 25319552 8579598 7887970 9359436 10825435 14702039 15489334 10330151 11747828 11713266 16087681 17525332 18984161 21081503 20075255 21719571 21292972 21347285
EMBL
BABH01024291
BABH01024292
BABH01024293
AGBW02008771
OWR52583.1
RSAL01000009
+ More
RVE53790.1 GAIX01004354 JAA88206.1 KQ459286 KPJ02032.1 ODYU01009556 SOQ54070.1 NWSH01001278 PCG71876.1 GDQN01011246 JAT79808.1 GDQN01010071 JAT80983.1 KZ288298 PBC28902.1 KQ434879 KZC09998.1 KK853129 KDR10972.1 KQ435794 KOX74043.1 KQ763839 OAD54701.1 OAD54702.1 GL443910 EFN61505.1 PYGN01001356 PSN35455.1 LBMM01001709 KMQ95856.1 KK107139 QOIP01000003 EZA57852.1 RLU24787.1 GAMT01005731 GAMS01009238 GAMR01000184 GAMQ01006551 JAB06130.1 JAB13898.1 JAB33748.1 JAB35300.1 AAQR03000925 AAQR03000926 AAQR03000927 AAQR03000928 AAQR03000929 AAQR03000930 AAQR03000931 AAQR03000932 AAQR03000933 AAQR03000934 JSUE03012989 AQIA01021516 CABD030080339 ADFV01111280 ADFV01111281 ADFV01111282 AJFE02024301 AJFE02024302 AJFE02024303 AHZZ02007182 NDHI03003437 PNJ51134.1 AACZ04016306 NBAG03000261 PNI56890.1 AP000609 AP002789 AP003680 MUZQ01000236 OWK54531.1 GDIP01078512 JAM25203.1 GDIP01110161 JAL93553.1 GDKW01000548 JAI56047.1 ACPB03006711 GL765434 EFZ16294.1 GL450678 EFN80649.1 MCFN01000216 OXB62431.1 GDIP01119659 GDIP01078514 GDIP01071719 JAL84055.1 GDIP01205750 JAJ17652.1 GDIQ01220869 GDIP01052620 JAK30856.1 JAM51095.1 LRGB01000642 KZS17342.1 AQIB01134068 AQIB01134069 AQIB01134070 AQIB01134071 ABGA01356287 ABGA01356288 ABGA01356289 ABGA01356290 ABGA01356291 ABGA01356292 PNJ51130.1 PNJ51133.1 JH331226 EHH62384.1 JU332866 JU474268 JV046158 CM001266 AFE76621.1 AFH31072.1 AFI36229.1 EHH23273.1 GABC01004969 GABF01002991 GABD01007684 GABE01001005 JAA06369.1 JAA19154.1 JAA25416.1 JAA43734.1 PNI56888.1 PNI56892.1 X91788 U17899 U53454 AF005422 AF026003 AF232708 AF232224 AF232225 AK315259 BT019907 BC119634 BC119635 CR860304 GDIP01249394 JAI74007.1 GDIQ01015326 JAN79411.1 GDIP01154000 JAJ69402.1 GL888624 EGI58873.1 KQ982074 KYQ60506.1 QRBI01000120 RMC07505.1 AHAT01003066 KQ979955 KYN18473.1 KQ981856 KYN34503.1 GL732596 EFX72742.1 KQ976490 KYM83353.1 ADTU01005314 ADTU01005315 NEVH01016296 PNF26001.1 PNF26002.1
RVE53790.1 GAIX01004354 JAA88206.1 KQ459286 KPJ02032.1 ODYU01009556 SOQ54070.1 NWSH01001278 PCG71876.1 GDQN01011246 JAT79808.1 GDQN01010071 JAT80983.1 KZ288298 PBC28902.1 KQ434879 KZC09998.1 KK853129 KDR10972.1 KQ435794 KOX74043.1 KQ763839 OAD54701.1 OAD54702.1 GL443910 EFN61505.1 PYGN01001356 PSN35455.1 LBMM01001709 KMQ95856.1 KK107139 QOIP01000003 EZA57852.1 RLU24787.1 GAMT01005731 GAMS01009238 GAMR01000184 GAMQ01006551 JAB06130.1 JAB13898.1 JAB33748.1 JAB35300.1 AAQR03000925 AAQR03000926 AAQR03000927 AAQR03000928 AAQR03000929 AAQR03000930 AAQR03000931 AAQR03000932 AAQR03000933 AAQR03000934 JSUE03012989 AQIA01021516 CABD030080339 ADFV01111280 ADFV01111281 ADFV01111282 AJFE02024301 AJFE02024302 AJFE02024303 AHZZ02007182 NDHI03003437 PNJ51134.1 AACZ04016306 NBAG03000261 PNI56890.1 AP000609 AP002789 AP003680 MUZQ01000236 OWK54531.1 GDIP01078512 JAM25203.1 GDIP01110161 JAL93553.1 GDKW01000548 JAI56047.1 ACPB03006711 GL765434 EFZ16294.1 GL450678 EFN80649.1 MCFN01000216 OXB62431.1 GDIP01119659 GDIP01078514 GDIP01071719 JAL84055.1 GDIP01205750 JAJ17652.1 GDIQ01220869 GDIP01052620 JAK30856.1 JAM51095.1 LRGB01000642 KZS17342.1 AQIB01134068 AQIB01134069 AQIB01134070 AQIB01134071 ABGA01356287 ABGA01356288 ABGA01356289 ABGA01356290 ABGA01356291 ABGA01356292 PNJ51130.1 PNJ51133.1 JH331226 EHH62384.1 JU332866 JU474268 JV046158 CM001266 AFE76621.1 AFH31072.1 AFI36229.1 EHH23273.1 GABC01004969 GABF01002991 GABD01007684 GABE01001005 JAA06369.1 JAA19154.1 JAA25416.1 JAA43734.1 PNI56888.1 PNI56892.1 X91788 U17899 U53454 AF005422 AF026003 AF232708 AF232224 AF232225 AK315259 BT019907 BC119634 BC119635 CR860304 GDIP01249394 JAI74007.1 GDIQ01015326 JAN79411.1 GDIP01154000 JAJ69402.1 GL888624 EGI58873.1 KQ982074 KYQ60506.1 QRBI01000120 RMC07505.1 AHAT01003066 KQ979955 KYN18473.1 KQ981856 KYN34503.1 GL732596 EFX72742.1 KQ976490 KYM83353.1 ADTU01005314 ADTU01005315 NEVH01016296 PNF26001.1 PNF26002.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000053268
UP000218220
UP000005203
+ More
UP000242457 UP000076502 UP000027135 UP000053105 UP000000311 UP000245037 UP000036403 UP000233040 UP000008225 UP000053097 UP000279307 UP000233020 UP000233160 UP000005225 UP000233120 UP000006718 UP000233180 UP000233100 UP000001519 UP000001073 UP000233140 UP000240080 UP000028761 UP000233200 UP000002277 UP000005640 UP000197619 UP000015103 UP000233080 UP000008237 UP000198323 UP000076858 UP000029965 UP000001595 UP000009130 UP000002358 UP000007755 UP000075809 UP000269221 UP000018468 UP000189704 UP000078492 UP000078541 UP000000305 UP000192224 UP000078540 UP000005205 UP000235965
UP000242457 UP000076502 UP000027135 UP000053105 UP000000311 UP000245037 UP000036403 UP000233040 UP000008225 UP000053097 UP000279307 UP000233020 UP000233160 UP000005225 UP000233120 UP000006718 UP000233180 UP000233100 UP000001519 UP000001073 UP000233140 UP000240080 UP000028761 UP000233200 UP000002277 UP000005640 UP000197619 UP000015103 UP000233080 UP000008237 UP000198323 UP000076858 UP000029965 UP000001595 UP000009130 UP000002358 UP000007755 UP000075809 UP000269221 UP000018468 UP000189704 UP000078492 UP000078541 UP000000305 UP000192224 UP000078540 UP000005205 UP000235965
Pfam
PF03517 Voldacs
Gene 3D
ProteinModelPortal
H9ISC8
A0A212FFT5
A0A3S2M953
S4P810
A0A0N1ID77
A0A2H1WLW8
+ More
A0A2A4JJC2 A0A1E1VYK6 A0A1E1W1Y9 A0A087ZX28 A0A2A3EC88 A0A154PFB1 A0A067R0J2 A0A0M9A1L2 A0A310SJX7 E2AYH0 A0A2P8XTY0 A0A0J7KZR6 A0A2K5R0P8 A0A2R8MJB4 A0A026WP68 A0A2K5R0P5 U3FEP2 A0A2K5BXI8 A0A2K6FA31 A0A2K6FA49 A0A2K5BXI5 H0X015 A0A2K6CNB8 A0A1D5QSS3 A0A2K6LHR9 A0A2K5VED0 G3QKQ8 A0A2I3HPW9 A0A2K5ZYD1 A0A2R9B7Q6 A0A2I3MUM9 A0A2K6RQ17 A0A2J8V0T9 A0A2I3RSE1 E9PJF4 A0A218ULT2 A0A0P5X2I9 A0A0P5USH3 A0A0P4VLQ5 T1HI80 A0A2K5KD94 E9ISZ4 E2BUH9 A0A226N4G7 A0A0P5U5B7 A0A0P5A0D7 A0A0P5YVA2 G1S432 A0A2K6RPY5 A0A2K6CN69 A0A165A9N0 A0A2K6LHN5 A0A2I2Z6E0 A0A2K5ZY78 A0A0D9QUZ6 A0A2R9B7R3 A0A096N4Y3 A0A2J8V0T8 G8F520 G7NEE5 K7DM64 P54105 Q5R719 A0A2K5KD78 A0A0P4X774 A0A0P6JL91 A0A0P5DMC0 A0A2K5C8U2 K7IZ42 F4X3S6 A0A151XJB0 A0A3M0K2M9 W5MHL2 A0A3Q0EBK3 A0A195E023 A0A195F210 E9H646 A0A2K6L244 A0A1W4YJE6 A0A195BFI1 A0A3Q0E7E2 A0A158P0L7 A0A2J7QBP2 A0A2J7QBP5
A0A2A4JJC2 A0A1E1VYK6 A0A1E1W1Y9 A0A087ZX28 A0A2A3EC88 A0A154PFB1 A0A067R0J2 A0A0M9A1L2 A0A310SJX7 E2AYH0 A0A2P8XTY0 A0A0J7KZR6 A0A2K5R0P8 A0A2R8MJB4 A0A026WP68 A0A2K5R0P5 U3FEP2 A0A2K5BXI8 A0A2K6FA31 A0A2K6FA49 A0A2K5BXI5 H0X015 A0A2K6CNB8 A0A1D5QSS3 A0A2K6LHR9 A0A2K5VED0 G3QKQ8 A0A2I3HPW9 A0A2K5ZYD1 A0A2R9B7Q6 A0A2I3MUM9 A0A2K6RQ17 A0A2J8V0T9 A0A2I3RSE1 E9PJF4 A0A218ULT2 A0A0P5X2I9 A0A0P5USH3 A0A0P4VLQ5 T1HI80 A0A2K5KD94 E9ISZ4 E2BUH9 A0A226N4G7 A0A0P5U5B7 A0A0P5A0D7 A0A0P5YVA2 G1S432 A0A2K6RPY5 A0A2K6CN69 A0A165A9N0 A0A2K6LHN5 A0A2I2Z6E0 A0A2K5ZY78 A0A0D9QUZ6 A0A2R9B7R3 A0A096N4Y3 A0A2J8V0T8 G8F520 G7NEE5 K7DM64 P54105 Q5R719 A0A2K5KD78 A0A0P4X774 A0A0P6JL91 A0A0P5DMC0 A0A2K5C8U2 K7IZ42 F4X3S6 A0A151XJB0 A0A3M0K2M9 W5MHL2 A0A3Q0EBK3 A0A195E023 A0A195F210 E9H646 A0A2K6L244 A0A1W4YJE6 A0A195BFI1 A0A3Q0E7E2 A0A158P0L7 A0A2J7QBP2 A0A2J7QBP5
PDB
1ZYI
E-value=1.95067e-12,
Score=171
Ontologies
KEGG
GO
PANTHER
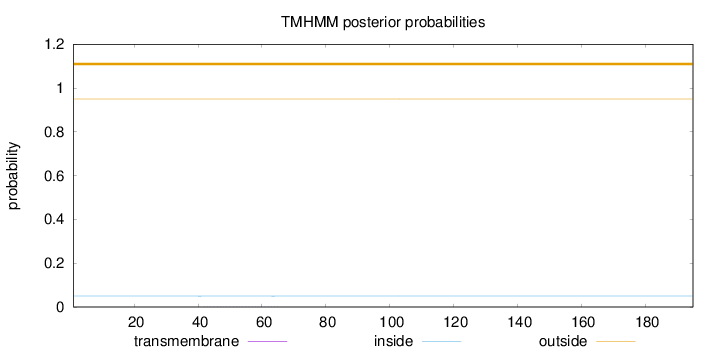
Topology
Subcellular location
Cytoplasm
Cytosol
Nucleus
Cytoskeleton
Cytosol
Nucleus
Cytoskeleton
Length:
195
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00794
Exp number, first 60 AAs:
0.00643
Total prob of N-in:
0.04992
outside
1 - 195
Population Genetic Test Statistics
Pi
237.915155
Theta
178.361644
Tajima's D
1.145319
CLR
0.184188
CSRT
0.693515324233788
Interpretation
Uncertain
