Gene
KWMTBOMO14479 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000074
Annotation
PREDICTED:_rRNA_2'-O-methyltransferase_fibrillarin_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.903
Sequence
CDS
ATGGGCAAACCTGAATTTAATGGCGGTCGCGGAGGAGGCGGCGGTCGCGGCGGTTTCCGTGGAGGTCGCGGCGGTGGCGGAGGCCGAGGTGGCTTCGGTGGCGGCCGTGGAGGCGGCGGTGGCGGCAAGTTCGAGAAGAGAGGCGGAGGCGGGGGCCGTGGCTTCGGAGGTAGAGGTGGTGGAGGCCGTGGTCGTGGTCGTGGTGGTCCCCCGAGCAGGGGAGGCTTCAAAGGCGGAAAACAAGTCATAATTGAACCACACAGGCATCCAGGAGTATTCATAGCTAGGGGCAAAGAAGATGCGTTGGTCACTAAAAATCTAGTGCCAGGATCAGAAGTTTATGGTGAAAAGAGAATATCAGTTGAGAACGAAGGTGATAAGGTGGAGTACAGAGTCTGGAATCCGTTCAGATCGAAATTGGCAGCTGCAATTATGGGTGGAGTGGATGCTATACACATGGCCCCTGGGTCAAGATAA
Protein
MGKPEFNGGRGGGGGRGGFRGGRGGGGGRGGFGGGRGGGGGGKFEKRGGGGGRGFGGRGGGGRGRGRGGPPSRGGFKGGKQVIIEPHRHPGVFIARGKEDALVTKNLVPGSEVYGEKRISVENEGDKVEYRVWNPFRSKLAAAIMGGVDAIHMAPGSR
Summary
Uniprot
ProteinModelPortal
PDB
2IPX
E-value=2.16545e-29,
Score=316
Ontologies
GO
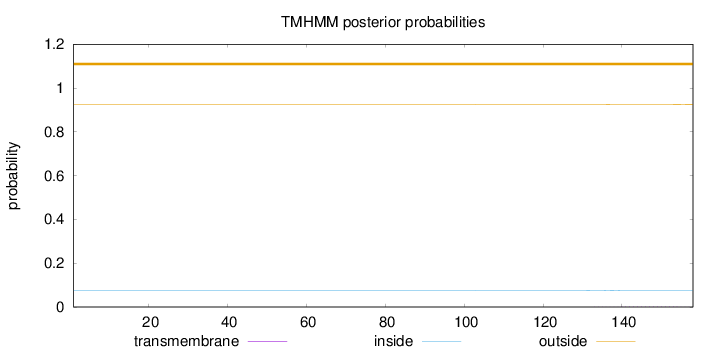
Topology
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0334
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.07571
outside
1 - 158
Population Genetic Test Statistics
Pi
327.215095
Theta
217.152325
Tajima's D
1.592273
CLR
0
CSRT
0.806759662016899
Interpretation
Possibly Positive selection
