Gene
KWMTBOMO14472
Pre Gene Modal
BGIBMGA000078
Annotation
PREDICTED:_transmembrane_channel-like_protein_7_[Bombyx_mori]
Full name
Transmembrane channel-like protein
Location in the cell
PlasmaMembrane Reliability : 4.937
Sequence
CDS
ATGTCAGGGGGTAACTCTAAGAACAAGGCGAAATCCAAACGCCATGAAGGTTGGGAGGAAGCCGGCGGGGAGTTCTACCAGGAACTCTACCCCGGGGGGGAACAGGAGCTGTTTGATAACCTCCAGAGAGCTGATGCAGCGAAACTGGCGACGCTACTACCATCAAAACAGGCTAGGAACACGACGACGGTCAAACGCGCCAGATCCCAGACTGATCGGCGCCAATCTACCTTCGCTAGGACCATGCAGAGCCGGGACATACATCTCTCGATGTTGCCAGACTTATCAGAAAATCTATCGAACGAAGAACGCTCGTGGGAGGAGATAATGCAGATAAAAGCGCTACCTGTCCCGATGAGTCAGAAGATCGAGCTTAAAGCTCGATTGCAGAACGCTACGAAACTTCGCCTCCAAGGTTTCGAGCAACTCCACTGGAGGCAGAGGAAGGTGTGGCACCGTTTCCGTATAGGCTGCACTGAGACGTTTAGCAAGCTGGAGCTCTGGCAGACCCCCATGCGTGAAATAGAAGGAAAATTCGGAACGGGAGTCGTCTCTTATTTCCTGTTCTTGAGGTGGCTCTTCCTGTTAAACTTCACAATATCGATACTGGTCATCGTGTTCCTGATCCTACCAAAGACATTGTTGACTGAGAGGGTTTACGAATGCGACGAGACGGAAGCGAATTCAACAGTTTGCTGTTCTATAGCGTATTTTGAGAAGAACACGACGAATTCAAATATATTTTTAGACGTTATCCAGGGCACTGGCTGGATGGAGAGAACCATACTCTTCTACGGAGTTTACACAGATCAAATATACACGTACTACGTAAATAGTTTATACCATACGAAACTGTTCTACAATATGCCAATGGCTTACATTTTGGTGCCGATTTCTTGGACTTTATTGTCCCTGATCGCGATCGTGAAGACGGCCGCCAGGGGCTTCAAGCAGAAGCTAATCGAAAGCGAGGGTCAGTTCTACAAGTACTGCAACCTCGTTTTTGGCGGTTGGGATTTTTGCATACACAACGACAGATCGGCGAGAATCAAACACCAAGCTCTGTACAATGAGATCAAGGGCTGCATCGAAGAGGAGAGATTCAAAGAAGAGAAACAGTCACGCAGCCGGGAGAGTCAGATATTGATGCACCTAAAAAGATTGTTTATTAACCTGATCGTGTTTGTGATACTCATAGCGTCCGGTTTTCTAATATACGTAGCCTTCAACTTCTCGACGGAGAGACTCAACGAAAGCATAGGATCTAACGAGAATCGAACCAGGACGATGCCGATAGTGTACACTACGATGTCTTTCAATCAATTGGAGAACGTACTAATGGAGTTTTTGCCGTTTATTTGCATTGTCGTACTGAATCTTATCATACCCGAGATTTTTAGTTACATGATCAGATACGAAGGCTACATGCCGCCACATGTTATCATTATAACGCTCCTAAGGACTGTGTTACTGAGGCTATCCTCGTTAGGCGTATTGTTAAGTCAAATCTATTTGTATATATCGACGAATGGCATCGTGTGCGCGAAGCATAACACGGATACAACGAAGTTAGAATGCTGGGAGACATACGTCGGTCAGCAGTTCTACAAGTTGATCTTGACCGATTTCGCCGTTCAATTCGTGACCACGTTCCTGATAAATTTGCCTAGGGCGTTTATGGCTCGTCACACGCGGAGCCGCTGCTTGAAGATGTTCGGCGAACAGGAATTCTATTTGCCTAAACACGTATTGGACATTGTTTACGTGCAGACCATAATTTGGATGGGTTCGTTCTTCTGTCCCTTTCTACCCATTATAGGATCTATGTTCTATTTCATGATATTCTATATAAAAAAGTTCACTTGCCTCGTAAACTGTACGCCCTCGCCGGTCGTGTATAAGGCGTCCAAATCGAAATCACTCTTCATGTCGGTTCTATTGCTCGGTTTCGTCATATCGATTCTGCCGGTAGCGTATTCGGTGGCGGAAATCTTGCCGTCTATAAACTGCGGTCCTTATCGAGGCTACACCACCGTGTGGGAATTCGTCATACAAACATTCAACAACTTTCCGTCGGTTATTAGAGAGGTCATATTCTTGTTAAGCACTGCAACGTTCGCGGTGCCGGCTTTCGCTGTGACATTATTCTTCTTGTATTATTACTGGGCCGTCGCGACAGCCAATAGGCACATGGTGGCGGTCCTTAAGAACCAACTGGTCTTAGAAGGCCACGATAAACAGTTTCTGTTGAACAGACTGAGCGCGTTTATACGGCAGCATCAGAAGAGGTGCGACAGAAGAAATAGGCCCAGTTTTGCGGACGATGACTCCTCTATAAGACAGAGTTCTAGATAG
Protein
MSGGNSKNKAKSKRHEGWEEAGGEFYQELYPGGEQELFDNLQRADAAKLATLLPSKQARNTTTVKRARSQTDRRQSTFARTMQSRDIHLSMLPDLSENLSNEERSWEEIMQIKALPVPMSQKIELKARLQNATKLRLQGFEQLHWRQRKVWHRFRIGCTETFSKLELWQTPMREIEGKFGTGVVSYFLFLRWLFLLNFTISILVIVFLILPKTLLTERVYECDETEANSTVCCSIAYFEKNTTNSNIFLDVIQGTGWMERTILFYGVYTDQIYTYYVNSLYHTKLFYNMPMAYILVPISWTLLSLIAIVKTAARGFKQKLIESEGQFYKYCNLVFGGWDFCIHNDRSARIKHQALYNEIKGCIEEERFKEEKQSRSRESQILMHLKRLFINLIVFVILIASGFLIYVAFNFSTERLNESIGSNENRTRTMPIVYTTMSFNQLENVLMEFLPFICIVVLNLIIPEIFSYMIRYEGYMPPHVIIITLLRTVLLRLSSLGVLLSQIYLYISTNGIVCAKHNTDTTKLECWETYVGQQFYKLILTDFAVQFVTTFLINLPRAFMARHTRSRCLKMFGEQEFYLPKHVLDIVYVQTIIWMGSFFCPFLPIIGSMFYFMIFYIKKFTCLVNCTPSPVVYKASKSKSLFMSVLLLGFVISILPVAYSVAEILPSINCGPYRGYTTVWEFVIQTFNNFPSVIREVIFLLSTATFAVPAFAVTLFFLYYYWAVATANRHMVAVLKNQLVLEGHDKQFLLNRLSAFIRQHQKRCDRRNRPSFADDDSSIRQSSR
Summary
Similarity
Belongs to the TMC family.
Uniprot
A0A2H1WPP0
A0A3S2P0G4
A0A2W1CMU3
H9IS52
A0A212FJD6
A0A2A4J8B3
+ More
A0A0L7L9P9 A0A1E1W9E7 S6D4Y2 A0A0C9RF80 A0A1Y1L4J7 A0A1W4XIW2 A0A026VXJ1 A0A1B0EY08 A0A2J7PFV7 Q17MQ2 E1ZVE5 K7J5H2 E2BC35 A0A067QMG9 A0A2A3E8M3 A0A1L8DET4 A0A1B6DP36 A0A0J7KZ66 D6WFV4 A0A0T6B3F6 A0A1S4H841 N6UCI0 A0A195BGG8 A0A158NBM8 E0W235 A0A195DS38 A0A151WLI1 A0A2J7PFX1 A0A2R7X2W0 A0A2P8Z0Q6 E9I9Z4 A0A195FE55 A0A195C6C4 A0A1B6IWP1 A0A0K8VRL6 F4X8H9 W8C034 A0A1B6LGE5 A0A0K8UN18 T1IBC9 A0A1L8DDS3 W8C5S9 A0A2H8TYL9 J9JYT7 A0A1B0CGV5 A0A0P4WRI4 A0A0P4WCK5 A0A1S4H998 A0A2S2P3L3 B0WFU4 Q7Z013 A0A182FUL2 A0A087Z986 T1GQ43 C3YWQ9 A0A084WI62 A0A224ZA29 A0A224ZAY6 A0A224ZAL8 A0A131Z6W6 A0A182KYC6 A0A182MJC3 A0A182YCG2 A0A182PLZ3 A0A182V6U2 A0A182RY45 A0A182K8V4 A0A1S3HSD2 A0A182HHV0 A0A1S3HV55 A0A3P9JU22 A0A182UBS3 A0A1S3HTR8 A0A182X7W9 A0A3P9JU15 A0A182WDA3 A0A3Q2DCH6 H2MDL0 A0A1L8EQ76 A0A3P9LKB2 V3ZD29 A0A1A8V601 A0A1A8I812 A0A1A8GP68 A0A1A8FQ54 A0A182ITY1 A0A131XPZ0 A0A1A8RAT4 A0A1S3HSD5 A0A1A8C5T0
A0A0L7L9P9 A0A1E1W9E7 S6D4Y2 A0A0C9RF80 A0A1Y1L4J7 A0A1W4XIW2 A0A026VXJ1 A0A1B0EY08 A0A2J7PFV7 Q17MQ2 E1ZVE5 K7J5H2 E2BC35 A0A067QMG9 A0A2A3E8M3 A0A1L8DET4 A0A1B6DP36 A0A0J7KZ66 D6WFV4 A0A0T6B3F6 A0A1S4H841 N6UCI0 A0A195BGG8 A0A158NBM8 E0W235 A0A195DS38 A0A151WLI1 A0A2J7PFX1 A0A2R7X2W0 A0A2P8Z0Q6 E9I9Z4 A0A195FE55 A0A195C6C4 A0A1B6IWP1 A0A0K8VRL6 F4X8H9 W8C034 A0A1B6LGE5 A0A0K8UN18 T1IBC9 A0A1L8DDS3 W8C5S9 A0A2H8TYL9 J9JYT7 A0A1B0CGV5 A0A0P4WRI4 A0A0P4WCK5 A0A1S4H998 A0A2S2P3L3 B0WFU4 Q7Z013 A0A182FUL2 A0A087Z986 T1GQ43 C3YWQ9 A0A084WI62 A0A224ZA29 A0A224ZAY6 A0A224ZAL8 A0A131Z6W6 A0A182KYC6 A0A182MJC3 A0A182YCG2 A0A182PLZ3 A0A182V6U2 A0A182RY45 A0A182K8V4 A0A1S3HSD2 A0A182HHV0 A0A1S3HV55 A0A3P9JU22 A0A182UBS3 A0A1S3HTR8 A0A182X7W9 A0A3P9JU15 A0A182WDA3 A0A3Q2DCH6 H2MDL0 A0A1L8EQ76 A0A3P9LKB2 V3ZD29 A0A1A8V601 A0A1A8I812 A0A1A8GP68 A0A1A8FQ54 A0A182ITY1 A0A131XPZ0 A0A1A8RAT4 A0A1S3HSD5 A0A1A8C5T0
Pubmed
28756777
19121390
22118469
26227816
23938757
28004739
+ More
24508170 30249741 17510324 20798317 20075255 24845553 18362917 19820115 12364791 23537049 21347285 20566863 29403074 21282665 21719571 24495485 12812529 14747013 17210077 20920257 18563158 24438588 28797301 26830274 20966253 25244985 17554307 27762356 23254933 28049606
24508170 30249741 17510324 20798317 20075255 24845553 18362917 19820115 12364791 23537049 21347285 20566863 29403074 21282665 21719571 24495485 12812529 14747013 17210077 20920257 18563158 24438588 28797301 26830274 20966253 25244985 17554307 27762356 23254933 28049606
EMBL
ODYU01010089
SOQ54946.1
RSAL01000009
RVE53797.1
NHMN01023779
PZC87381.1
+ More
BABH01024272 BABH01024273 AGBW02008275 OWR53853.1 NWSH01002714 PCG67633.1 JTDY01002102 KOB72114.1 GDQN01007485 JAT83569.1 HF586479 CCQ71377.1 GBYB01007065 JAG76832.1 GEZM01067948 JAV67310.1 KK107638 QOIP01000004 EZA48390.1 RLU23213.1 AJVK01015212 AJVK01015213 NEVH01025648 PNF15216.1 CH477204 EAT47970.1 GL434492 EFN74855.1 GL447247 EFN86760.1 KK853149 KDR10638.1 KZ288324 PBC28075.1 GFDF01009112 JAV04972.1 GEDC01009866 JAS27432.1 LBMM01001902 KMQ95578.1 KQ971328 EFA00942.1 LJIG01016080 KRT81675.1 AAAB01008986 APGK01034495 KB740914 KB632005 ENN78341.1 ERL87924.1 KQ976500 KYM83245.1 ADTU01011227 DS235874 EEB19691.1 KQ980581 KYN15334.1 KQ982981 KYQ48545.1 PNF15217.1 KK856629 PTY26139.1 PYGN01000252 PSN50086.1 GL761864 EFZ22606.1 KQ981673 KYN38304.1 KQ978219 KYM96397.1 GECU01016368 JAS91338.1 GDHF01011109 JAI41205.1 GL888932 EGI57248.1 GAMC01001548 JAC05008.1 GEBQ01017333 JAT22644.1 GDHF01024594 JAI27720.1 ACPB03006691 GFDF01009594 JAV04490.1 GAMC01001547 JAC05009.1 GFXV01006817 MBW18622.1 ABLF02035184 AJWK01011608 GDRN01068408 JAI64196.1 GDRN01068409 GDRN01068407 JAI64195.1 GGMR01011189 MBY23808.1 DS231920 EDS26438.1 AY263177 AAP78792.1 EAA00457.3 CAQQ02064822 CAQQ02064823 CAQQ02064824 CAQQ02064825 CAQQ02064826 CAQQ02064827 GG666561 EEN55303.1 ATLV01023914 KE525347 KFB49906.1 GFPF01012777 MAA23923.1 GFPF01012776 MAA23922.1 GFPF01012778 MAA23924.1 GEDV01001473 JAP87084.1 AXCM01007794 APCN01002444 CM004483 OCT61504.1 KB202619 ESO89008.1 HAEJ01015682 SBS56139.1 HAED01007444 SBQ93656.1 HAEC01005558 SBQ73635.1 HAEB01014451 SBQ60978.1 GEFH01000950 JAP67631.1 HAEH01015291 SBS02364.1 HADZ01011008 SBP74949.1
BABH01024272 BABH01024273 AGBW02008275 OWR53853.1 NWSH01002714 PCG67633.1 JTDY01002102 KOB72114.1 GDQN01007485 JAT83569.1 HF586479 CCQ71377.1 GBYB01007065 JAG76832.1 GEZM01067948 JAV67310.1 KK107638 QOIP01000004 EZA48390.1 RLU23213.1 AJVK01015212 AJVK01015213 NEVH01025648 PNF15216.1 CH477204 EAT47970.1 GL434492 EFN74855.1 GL447247 EFN86760.1 KK853149 KDR10638.1 KZ288324 PBC28075.1 GFDF01009112 JAV04972.1 GEDC01009866 JAS27432.1 LBMM01001902 KMQ95578.1 KQ971328 EFA00942.1 LJIG01016080 KRT81675.1 AAAB01008986 APGK01034495 KB740914 KB632005 ENN78341.1 ERL87924.1 KQ976500 KYM83245.1 ADTU01011227 DS235874 EEB19691.1 KQ980581 KYN15334.1 KQ982981 KYQ48545.1 PNF15217.1 KK856629 PTY26139.1 PYGN01000252 PSN50086.1 GL761864 EFZ22606.1 KQ981673 KYN38304.1 KQ978219 KYM96397.1 GECU01016368 JAS91338.1 GDHF01011109 JAI41205.1 GL888932 EGI57248.1 GAMC01001548 JAC05008.1 GEBQ01017333 JAT22644.1 GDHF01024594 JAI27720.1 ACPB03006691 GFDF01009594 JAV04490.1 GAMC01001547 JAC05009.1 GFXV01006817 MBW18622.1 ABLF02035184 AJWK01011608 GDRN01068408 JAI64196.1 GDRN01068409 GDRN01068407 JAI64195.1 GGMR01011189 MBY23808.1 DS231920 EDS26438.1 AY263177 AAP78792.1 EAA00457.3 CAQQ02064822 CAQQ02064823 CAQQ02064824 CAQQ02064825 CAQQ02064826 CAQQ02064827 GG666561 EEN55303.1 ATLV01023914 KE525347 KFB49906.1 GFPF01012777 MAA23923.1 GFPF01012776 MAA23922.1 GFPF01012778 MAA23924.1 GEDV01001473 JAP87084.1 AXCM01007794 APCN01002444 CM004483 OCT61504.1 KB202619 ESO89008.1 HAEJ01015682 SBS56139.1 HAED01007444 SBQ93656.1 HAEC01005558 SBQ73635.1 HAEB01014451 SBQ60978.1 GEFH01000950 JAP67631.1 HAEH01015291 SBS02364.1 HADZ01011008 SBP74949.1
Proteomes
UP000283053
UP000005204
UP000007151
UP000218220
UP000037510
UP000192223
+ More
UP000053097 UP000279307 UP000092462 UP000235965 UP000008820 UP000000311 UP000002358 UP000008237 UP000027135 UP000242457 UP000036403 UP000007266 UP000019118 UP000030742 UP000078540 UP000005205 UP000009046 UP000078492 UP000075809 UP000245037 UP000078541 UP000078542 UP000007755 UP000015103 UP000007819 UP000092461 UP000002320 UP000007062 UP000069272 UP000015102 UP000001554 UP000030765 UP000075882 UP000075883 UP000076408 UP000075885 UP000075903 UP000075900 UP000075881 UP000085678 UP000075840 UP000265200 UP000075902 UP000076407 UP000075920 UP000265020 UP000001038 UP000186698 UP000265180 UP000030746 UP000075880
UP000053097 UP000279307 UP000092462 UP000235965 UP000008820 UP000000311 UP000002358 UP000008237 UP000027135 UP000242457 UP000036403 UP000007266 UP000019118 UP000030742 UP000078540 UP000005205 UP000009046 UP000078492 UP000075809 UP000245037 UP000078541 UP000078542 UP000007755 UP000015103 UP000007819 UP000092461 UP000002320 UP000007062 UP000069272 UP000015102 UP000001554 UP000030765 UP000075882 UP000075883 UP000076408 UP000075885 UP000075903 UP000075900 UP000075881 UP000085678 UP000075840 UP000265200 UP000075902 UP000076407 UP000075920 UP000265020 UP000001038 UP000186698 UP000265180 UP000030746 UP000075880
PRIDE
ProteinModelPortal
A0A2H1WPP0
A0A3S2P0G4
A0A2W1CMU3
H9IS52
A0A212FJD6
A0A2A4J8B3
+ More
A0A0L7L9P9 A0A1E1W9E7 S6D4Y2 A0A0C9RF80 A0A1Y1L4J7 A0A1W4XIW2 A0A026VXJ1 A0A1B0EY08 A0A2J7PFV7 Q17MQ2 E1ZVE5 K7J5H2 E2BC35 A0A067QMG9 A0A2A3E8M3 A0A1L8DET4 A0A1B6DP36 A0A0J7KZ66 D6WFV4 A0A0T6B3F6 A0A1S4H841 N6UCI0 A0A195BGG8 A0A158NBM8 E0W235 A0A195DS38 A0A151WLI1 A0A2J7PFX1 A0A2R7X2W0 A0A2P8Z0Q6 E9I9Z4 A0A195FE55 A0A195C6C4 A0A1B6IWP1 A0A0K8VRL6 F4X8H9 W8C034 A0A1B6LGE5 A0A0K8UN18 T1IBC9 A0A1L8DDS3 W8C5S9 A0A2H8TYL9 J9JYT7 A0A1B0CGV5 A0A0P4WRI4 A0A0P4WCK5 A0A1S4H998 A0A2S2P3L3 B0WFU4 Q7Z013 A0A182FUL2 A0A087Z986 T1GQ43 C3YWQ9 A0A084WI62 A0A224ZA29 A0A224ZAY6 A0A224ZAL8 A0A131Z6W6 A0A182KYC6 A0A182MJC3 A0A182YCG2 A0A182PLZ3 A0A182V6U2 A0A182RY45 A0A182K8V4 A0A1S3HSD2 A0A182HHV0 A0A1S3HV55 A0A3P9JU22 A0A182UBS3 A0A1S3HTR8 A0A182X7W9 A0A3P9JU15 A0A182WDA3 A0A3Q2DCH6 H2MDL0 A0A1L8EQ76 A0A3P9LKB2 V3ZD29 A0A1A8V601 A0A1A8I812 A0A1A8GP68 A0A1A8FQ54 A0A182ITY1 A0A131XPZ0 A0A1A8RAT4 A0A1S3HSD5 A0A1A8C5T0
A0A0L7L9P9 A0A1E1W9E7 S6D4Y2 A0A0C9RF80 A0A1Y1L4J7 A0A1W4XIW2 A0A026VXJ1 A0A1B0EY08 A0A2J7PFV7 Q17MQ2 E1ZVE5 K7J5H2 E2BC35 A0A067QMG9 A0A2A3E8M3 A0A1L8DET4 A0A1B6DP36 A0A0J7KZ66 D6WFV4 A0A0T6B3F6 A0A1S4H841 N6UCI0 A0A195BGG8 A0A158NBM8 E0W235 A0A195DS38 A0A151WLI1 A0A2J7PFX1 A0A2R7X2W0 A0A2P8Z0Q6 E9I9Z4 A0A195FE55 A0A195C6C4 A0A1B6IWP1 A0A0K8VRL6 F4X8H9 W8C034 A0A1B6LGE5 A0A0K8UN18 T1IBC9 A0A1L8DDS3 W8C5S9 A0A2H8TYL9 J9JYT7 A0A1B0CGV5 A0A0P4WRI4 A0A0P4WCK5 A0A1S4H998 A0A2S2P3L3 B0WFU4 Q7Z013 A0A182FUL2 A0A087Z986 T1GQ43 C3YWQ9 A0A084WI62 A0A224ZA29 A0A224ZAY6 A0A224ZAL8 A0A131Z6W6 A0A182KYC6 A0A182MJC3 A0A182YCG2 A0A182PLZ3 A0A182V6U2 A0A182RY45 A0A182K8V4 A0A1S3HSD2 A0A182HHV0 A0A1S3HV55 A0A3P9JU22 A0A182UBS3 A0A1S3HTR8 A0A182X7W9 A0A3P9JU15 A0A182WDA3 A0A3Q2DCH6 H2MDL0 A0A1L8EQ76 A0A3P9LKB2 V3ZD29 A0A1A8V601 A0A1A8I812 A0A1A8GP68 A0A1A8FQ54 A0A182ITY1 A0A131XPZ0 A0A1A8RAT4 A0A1S3HSD5 A0A1A8C5T0
Ontologies
KEGG
GO
PANTHER
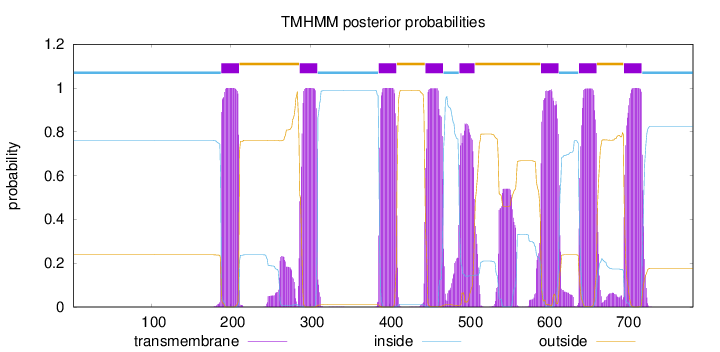
Topology
Subcellular location
Membrane
Length:
784
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
196.57592
Exp number, first 60 AAs:
0
Total prob of N-in:
0.76163
inside
1 - 187
TMhelix
188 - 210
outside
211 - 286
TMhelix
287 - 309
inside
310 - 386
TMhelix
387 - 409
outside
410 - 445
TMhelix
446 - 468
inside
469 - 488
TMhelix
489 - 508
outside
509 - 591
TMhelix
592 - 614
inside
615 - 639
TMhelix
640 - 662
outside
663 - 696
TMhelix
697 - 719
inside
720 - 784
Population Genetic Test Statistics
Pi
253.959441
Theta
167.11104
Tajima's D
1.06488
CLR
0.12158
CSRT
0.67621618919054
Interpretation
Uncertain
