Gene
KWMTBOMO14470
Pre Gene Modal
BGIBMGA000149
Annotation
PREDICTED:_tubulin-specific_chaperone_E_[Papilio_polytes]
Full name
Lactoylglutathione lyase
+ More
Tubulin-specific chaperone E
Tubulin-specific chaperone E
Alternative Name
Glyoxalase I
Tubulin-folding cofactor E
Tubulin-folding cofactor E
Location in the cell
Cytoplasmic Reliability : 2.365
Sequence
CDS
ATGGTGACGTTCGGCGTATTTCAAACTTTAGAGTCCCAAGTGGAATTCGGACGCTCGGAAGAGGAGTTGAGGAGAGTCACACATACACTACGCGTTTCCTCGGTAGAGAATATCGCTGGAGACTTGGCTTTCGACATAAGAAAGTATATTGGTGAGGTTCAGGGCTACAAGGGATTATGGTACGGCATAGAATGGGACAACGAGTCCAGAGGCAAGCACGACGGGTGCTTAGATGGCATCCAGTACTTCAAGACTTCCAAACCTAGTGCCGGTTCTTTTGTTCGTCCAAACAAGATCATCCCATCGAAGACTTGCGCGGAAGCCATAAGACAATATTATGGAGATCCGGAGGACGAAACATTAGCTGCTCACCGCAGAACCATCATCAATGAATGGAAGAGGGAAATGGGAGCGCCTTTCATCGAAATGGTTGGTTTTGAAAAAATACACCAAAAACAGAAATTTGATCGCTTACTAGAAGTATGCGTTCACGATCAGAATGTATCAAGAGCTGGTGATGTGTCTTCTCTATGTCCCAACGTGAGAAGCCTCGACGTCTCACGTAACCTCTTCTCCAACTGGCGTGAGGTCATACAACTGTCTGCTCAACTACCGGACTTGAGAGATTTGGATGTTAGCAAGAACCGTATGCTCATCGATGCGCCCACAGAAATACTTGCACAACTTTCAATCAACTTTTCCAAGCTTGAAAAGATCAATCTTTCTGTCTGCGATTATACATGGACGGACATATTGAAATTATCACATTTATGGCCTAAGATACATGAGATAATAGCTGCCTACAATCGTATAGATGAAATCGAACCACCGCTTGTAACTCTGAGGAGTTTATCTGTGCTGAGACTGGATGGAAACCCGATCACCTCTTGGAGTGAAGTGATGAATCTTGGTGCTCTCAATCTGAAGGTTTTGAGTTTGAACGACTGTCTCATTGCGGAGATTAGGTTCGGTGATGATTGGAATCAGAGAGTTGAGACATTTAAAAATTTGGAGATTTTGTTTTTGAATCGAAATAGGATTAATGATTGGCGTTCCATAAGTGAATTAAACAAACTTAGCTCGTTGAAAAAGCTTTATTTCCTCAAGAATCCGATACAAACCACCGAAGACTACGACACAGGATCGCAGCTGGTTGTCGCTAAAATTGGCACTTTACAGGAATTGAATGGCTCAATAATAACCGGGGAGTTGCGTAGAGGCGCTGAATACGATTACCTGAAGCGTTACGGAGGAGAATGGAAGGTCGCTCAGCTCGATTTTGAGGCGAAAGCGGCCTTCGATGCCGAACATTGTCGCTTTGAGGAGCTCATCCGTAAATACGGAATACCGGAAGACAGCCTCTTGGTCAAGCTGCCGAAAATAACGACGCTGACATCGCAACTTTTAGAAGTAACATTGAGAGACGAAAACGGAAAATCGTTTAAGAAAAAGTTCCCATCTACGATGGCAGTCCAGAAGCTGATCACACTGTCGCAGAGATTGTTCTCGAGAGCCGGCAACACCGGTACACCTCGACTTTACGTACTAGACGAACAGATGGATGGGGCTGAAATATATCTCGACAATTCTATGAAGGATCTCGCTTATTATTCGATTAAAAATGGCGATGTTATTCTAGTTAAATTCAGATAG
Protein
MVTFGVFQTLESQVEFGRSEEELRRVTHTLRVSSVENIAGDLAFDIRKYIGEVQGYKGLWYGIEWDNESRGKHDGCLDGIQYFKTSKPSAGSFVRPNKIIPSKTCAEAIRQYYGDPEDETLAAHRRTIINEWKREMGAPFIEMVGFEKIHQKQKFDRLLEVCVHDQNVSRAGDVSSLCPNVRSLDVSRNLFSNWREVIQLSAQLPDLRDLDVSKNRMLIDAPTEILAQLSINFSKLEKINLSVCDYTWTDILKLSHLWPKIHEIIAAYNRIDEIEPPLVTLRSLSVLRLDGNPITSWSEVMNLGALNLKVLSLNDCLIAEIRFGDDWNQRVETFKNLEILFLNRNRINDWRSISELNKLSSLKKLYFLKNPIQTTEDYDTGSQLVVAKIGTLQELNGSIITGELRRGAEYDYLKRYGGEWKVAQLDFEAKAAFDAEHCRFEELIRKYGIPEDSLLVKLPKITTLTSQLLEVTLRDENGKSFKKKFPSTMAVQKLITLSQRLFSRAGNTGTPRLYVLDEQMDGAEIYLDNSMKDLAYYSIKNGDVILVKFR
Summary
Description
Catalyzes the conversion of hemimercaptal, formed from methylglyoxal and glutathione, to S-lactoylglutathione.
Tubulin-folding protein; involved in the second step of the tubulin folding pathway and in the regulation of tubulin heterodimer dissociation (PubMed:12389029, PubMed:17184771). Required for correct organization of microtubule cytoskeleton and mitotic splindle, and maintenance of the neuronal microtubule network (By similarity).
Tubulin-folding protein; involved in the second step of the tubulin folding pathway and in the regulation of tubulin heterodimer dissociation (PubMed:12389029, PubMed:17184771). Required for correct organization of microtubule cytoskeleton and mitotic splindle, and maintenance of the neuronal microtubule network (By similarity).
Catalytic Activity
(R)-S-lactoylglutathione = glutathione + methylglyoxal
Cofactor
Zn(2+)
Subunit
Supercomplex made of cofactors A to E. Cofactors A and D function by capturing and stabilizing tubulin in a quasi-native conformation. Cofactor E binds to the cofactor D-tubulin complex; interaction with cofactor C then causes the release of tubulin polypeptides that are committed to the native state.
Supercomplex made of cofactors A to E. Cofactors A and D function by capturing and stabilizing tubulin in a quasi-native conformation. Cofactor E binds to the cofactor D-tubulin complex; interaction with cofactor C then causes the release of tubulin polypeptides that are committed to the native state. Cofactors B and E can form a heterodimer which binds to alpha-tubulin and enhances their ability to dissociate tubulin heterodimers. Interacts with TBCD (By similarity).
Supercomplex made of cofactors A to E. Cofactors A and D function by capturing and stabilizing tubulin in a quasi-native conformation. Cofactor E binds to the cofactor D-tubulin complex; interaction with cofactor C then causes the release of tubulin polypeptides that are committed to the native state. Cofactors B and E can form a heterodimer which binds to alpha-tubulin and enhances their ability to dissociate tubulin heterodimers. Interacts with TBCD (By similarity).
Similarity
Belongs to the glyoxalase I family.
Belongs to the TBCE family.
Belongs to the TBCE family.
Keywords
Chaperone
Cytoplasm
Cytoskeleton
Leucine-rich repeat
Repeat
3D-structure
Acetylation
Complete proteome
Phosphoprotein
Polymorphism
Reference proteome
Feature
chain Tubulin-specific chaperone E
Uniprot
H9ISC2
A0A2H1WCF3
A0A2A4JYI3
A0A0N1I4V6
A0A2W1BFL5
A0A212FJE5
+ More
A0A212EWZ1 A0A194RJW2 A0A158N911 D6WI91 A0A1Y1L5Y1 A0A1Y1L4R6 A0A195BUY8 K7IS98 A0A2C9LD34 A0A154P8U1 A0A0N0BFS8 A0A151WNS7 A0A088ALP0 A0A1W4X421 A0A151J375 A0A0L7QRM2 A0A2A3ETV0 A0A2J7QV38 A0A0J7L128 A0A151K114 E0VEM4 E2BBM1 A0A151IBY3 E2AAG3 A0A232EUB5 A0A310SAK1 A0A1B6EEL5 A0A2J7QV34 A0A0P4XVY7 A0A0P4YSW9 A0A0P6IKA5 V4AGM2 A0A026W415 A0A0P5ZMU4 A0A023F7S1 A0A067RA53 A0A0P6AAN6 A0A1S3IDI9 K7G0W2 A0A1S3ICD7 A0A3B3SA87 A0A0P6DXF3 N6TA25 A0A151M2W4 A0A1B6I484 A0A224XMS2 A0A2D4HA23 A0A0A9Y8G9 A0A0P5CE79 A0A2D4HAA5 A0A0P4VTZ4 A0A1U7RAE4 A0A3B4DXG5 R4GAP8 M3XIN9 A0A060Y2L9 A0A1W7RC53 R4G3X9 V9KB66 A0A1S3L6Y8 R0LCY4 A0A210Q0C9 W5LPV5 T1J621 A0A3P8YIC3 J3S5D9 T1D7I4 A0A0F7Z1T7 W5UCG8 U3IDV7 A0A093RIJ3 A0A1I8NRU6 A0A091G566 H0ZJ32 G3WPP0 A0A1W4VMM1 A0A0P5YR06 A0A1W4VMD9 A0A1L8G8N3 Q5U508 Q8CIV8 W5NG15 A0A091Q4U8 A0A0V0G618 A0A218VAM3 A0A2I0UR94 I3MQ76 B4NYV6 A0A1W4ZSA6 A0A3Q3MKF1 A0A0K8VJ60
A0A212EWZ1 A0A194RJW2 A0A158N911 D6WI91 A0A1Y1L5Y1 A0A1Y1L4R6 A0A195BUY8 K7IS98 A0A2C9LD34 A0A154P8U1 A0A0N0BFS8 A0A151WNS7 A0A088ALP0 A0A1W4X421 A0A151J375 A0A0L7QRM2 A0A2A3ETV0 A0A2J7QV38 A0A0J7L128 A0A151K114 E0VEM4 E2BBM1 A0A151IBY3 E2AAG3 A0A232EUB5 A0A310SAK1 A0A1B6EEL5 A0A2J7QV34 A0A0P4XVY7 A0A0P4YSW9 A0A0P6IKA5 V4AGM2 A0A026W415 A0A0P5ZMU4 A0A023F7S1 A0A067RA53 A0A0P6AAN6 A0A1S3IDI9 K7G0W2 A0A1S3ICD7 A0A3B3SA87 A0A0P6DXF3 N6TA25 A0A151M2W4 A0A1B6I484 A0A224XMS2 A0A2D4HA23 A0A0A9Y8G9 A0A0P5CE79 A0A2D4HAA5 A0A0P4VTZ4 A0A1U7RAE4 A0A3B4DXG5 R4GAP8 M3XIN9 A0A060Y2L9 A0A1W7RC53 R4G3X9 V9KB66 A0A1S3L6Y8 R0LCY4 A0A210Q0C9 W5LPV5 T1J621 A0A3P8YIC3 J3S5D9 T1D7I4 A0A0F7Z1T7 W5UCG8 U3IDV7 A0A093RIJ3 A0A1I8NRU6 A0A091G566 H0ZJ32 G3WPP0 A0A1W4VMM1 A0A0P5YR06 A0A1W4VMD9 A0A1L8G8N3 Q5U508 Q8CIV8 W5NG15 A0A091Q4U8 A0A0V0G618 A0A218VAM3 A0A2I0UR94 I3MQ76 B4NYV6 A0A1W4ZSA6 A0A3Q3MKF1 A0A0K8VJ60
EC Number
4.4.1.5
Pubmed
19121390
26354079
28756777
22118469
21347285
18362917
+ More
19820115 28004739 20075255 15562597 20566863 20798317 28648823 23254933 24508170 30249741 25474469 24845553 17381049 29240929 23537049 22293439 25401762 26823975 27129103 9215903 24755649 26358130 24402279 28812685 25329095 25069045 23025625 23758969 23127152 23749191 20360741 21709235 27762356 12389029 16141072 15489334 17184771 21183079 17994087 17550304
19820115 28004739 20075255 15562597 20566863 20798317 28648823 23254933 24508170 30249741 25474469 24845553 17381049 29240929 23537049 22293439 25401762 26823975 27129103 9215903 24755649 26358130 24402279 28812685 25329095 25069045 23025625 23758969 23127152 23749191 20360741 21709235 27762356 12389029 16141072 15489334 17184771 21183079 17994087 17550304
EMBL
BABH01024270
BABH01024271
ODYU01007715
SOQ50743.1
NWSH01000390
PCG76738.1
+ More
KQ459286 KPJ02021.1 KZ150604 PZC70463.1 AGBW02008275 OWR53855.1 AGBW02011887 OWR45967.1 KQ460118 KPJ17724.1 ADTU01009023 KQ971321 EFA01432.2 GEZM01068371 JAV66956.1 GEZM01068372 JAV66955.1 KQ976405 KYM91895.1 KQ434827 KZC07638.1 KQ435794 KOX73784.1 KQ982907 KYQ49457.1 KQ980290 KYN16910.1 KQ414775 KOC61278.1 KZ288189 PBC34569.1 NEVH01010479 PNF32445.1 LBMM01001332 KMQ96492.1 KQ981229 KYN44335.1 DS235093 EEB11830.1 GL447128 EFN86898.1 KQ978095 KYM97018.1 GL438125 EFN69564.1 NNAY01002165 OXU21896.1 KQ762190 OAD56160.1 GEDC01020706 GEDC01000958 JAS16592.1 JAS36340.1 PNF32444.1 GDIP01238245 JAI85156.1 GDIP01227634 JAI95767.1 GDIQ01020424 JAN74313.1 KB201847 ESO94310.1 KK107459 QOIP01000004 EZA50346.1 RLU23316.1 GDIP01042597 JAM61118.1 GBBI01001458 JAC17254.1 KK852595 KDR20629.1 GDIP01033202 JAM70513.1 AGCU01078981 AGCU01078982 AGCU01078983 AGCU01078984 AGCU01078985 AGCU01078986 AGCU01078987 GDIQ01070854 JAN23883.1 APGK01038391 APGK01038392 KB740954 KB632338 ENN77099.1 ERL92904.1 AKHW03006780 KYO18826.1 GECU01025960 JAS81746.1 GFTR01006983 JAW09443.1 IACK01016516 LAA68838.1 GBHO01016211 GBRD01015944 GDHC01003000 JAG27393.1 JAG49882.1 JAQ15629.1 GDIP01171832 JAJ51570.1 IACK01016515 IACK01016517 LAA68836.1 GDKW01001113 JAI55482.1 AFYH01046657 AFYH01046658 AFYH01046659 AFYH01046660 AFYH01046661 AFYH01046662 AFYH01046663 AFYH01046664 FR906223 CDQ83639.1 GDAY02002745 JAV48722.1 ACPB03019875 GAHY01001276 JAA76234.1 JW862515 AFO95032.1 KB743543 EOA98157.1 NEDP02005311 OWF42186.1 JH431869 JU176344 AFJ51867.1 GAAZ01002739 JAA95204.1 GBEX01003735 JAI10825.1 JT405921 AHH37382.1 ADON01100450 KL225298 KFW70709.1 KL447787 KFO77088.1 ABQF01014603 AEFK01160097 AEFK01160098 AEFK01160099 AEFK01160100 AEFK01160101 AEFK01160102 GDIP01054864 JAM48851.1 CM004474 OCT80064.1 BC084879 AY082332 AK167383 BC050206 AHAT01001606 KK655550 KFQ04669.1 GECL01002638 JAP03486.1 MUZQ01000016 OWK63097.1 KZ505650 PKU48569.1 AGTP01017245 AGTP01017246 AGTP01017247 AGTP01017248 CM000157 EDW89807.1 GDHF01025707 GDHF01013390 JAI26607.1 JAI38924.1
KQ459286 KPJ02021.1 KZ150604 PZC70463.1 AGBW02008275 OWR53855.1 AGBW02011887 OWR45967.1 KQ460118 KPJ17724.1 ADTU01009023 KQ971321 EFA01432.2 GEZM01068371 JAV66956.1 GEZM01068372 JAV66955.1 KQ976405 KYM91895.1 KQ434827 KZC07638.1 KQ435794 KOX73784.1 KQ982907 KYQ49457.1 KQ980290 KYN16910.1 KQ414775 KOC61278.1 KZ288189 PBC34569.1 NEVH01010479 PNF32445.1 LBMM01001332 KMQ96492.1 KQ981229 KYN44335.1 DS235093 EEB11830.1 GL447128 EFN86898.1 KQ978095 KYM97018.1 GL438125 EFN69564.1 NNAY01002165 OXU21896.1 KQ762190 OAD56160.1 GEDC01020706 GEDC01000958 JAS16592.1 JAS36340.1 PNF32444.1 GDIP01238245 JAI85156.1 GDIP01227634 JAI95767.1 GDIQ01020424 JAN74313.1 KB201847 ESO94310.1 KK107459 QOIP01000004 EZA50346.1 RLU23316.1 GDIP01042597 JAM61118.1 GBBI01001458 JAC17254.1 KK852595 KDR20629.1 GDIP01033202 JAM70513.1 AGCU01078981 AGCU01078982 AGCU01078983 AGCU01078984 AGCU01078985 AGCU01078986 AGCU01078987 GDIQ01070854 JAN23883.1 APGK01038391 APGK01038392 KB740954 KB632338 ENN77099.1 ERL92904.1 AKHW03006780 KYO18826.1 GECU01025960 JAS81746.1 GFTR01006983 JAW09443.1 IACK01016516 LAA68838.1 GBHO01016211 GBRD01015944 GDHC01003000 JAG27393.1 JAG49882.1 JAQ15629.1 GDIP01171832 JAJ51570.1 IACK01016515 IACK01016517 LAA68836.1 GDKW01001113 JAI55482.1 AFYH01046657 AFYH01046658 AFYH01046659 AFYH01046660 AFYH01046661 AFYH01046662 AFYH01046663 AFYH01046664 FR906223 CDQ83639.1 GDAY02002745 JAV48722.1 ACPB03019875 GAHY01001276 JAA76234.1 JW862515 AFO95032.1 KB743543 EOA98157.1 NEDP02005311 OWF42186.1 JH431869 JU176344 AFJ51867.1 GAAZ01002739 JAA95204.1 GBEX01003735 JAI10825.1 JT405921 AHH37382.1 ADON01100450 KL225298 KFW70709.1 KL447787 KFO77088.1 ABQF01014603 AEFK01160097 AEFK01160098 AEFK01160099 AEFK01160100 AEFK01160101 AEFK01160102 GDIP01054864 JAM48851.1 CM004474 OCT80064.1 BC084879 AY082332 AK167383 BC050206 AHAT01001606 KK655550 KFQ04669.1 GECL01002638 JAP03486.1 MUZQ01000016 OWK63097.1 KZ505650 PKU48569.1 AGTP01017245 AGTP01017246 AGTP01017247 AGTP01017248 CM000157 EDW89807.1 GDHF01025707 GDHF01013390 JAI26607.1 JAI38924.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000005205
+ More
UP000007266 UP000078540 UP000002358 UP000076420 UP000076502 UP000053105 UP000075809 UP000005203 UP000192223 UP000078492 UP000053825 UP000242457 UP000235965 UP000036403 UP000078541 UP000009046 UP000008237 UP000078542 UP000000311 UP000215335 UP000030746 UP000053097 UP000279307 UP000027135 UP000085678 UP000007267 UP000261540 UP000019118 UP000030742 UP000050525 UP000189705 UP000261440 UP000001646 UP000008672 UP000193380 UP000015103 UP000087266 UP000242188 UP000018467 UP000265140 UP000221080 UP000016666 UP000054081 UP000095300 UP000053760 UP000007754 UP000007648 UP000192221 UP000186698 UP000000589 UP000018468 UP000197619 UP000005215 UP000002282 UP000192224 UP000261640
UP000007266 UP000078540 UP000002358 UP000076420 UP000076502 UP000053105 UP000075809 UP000005203 UP000192223 UP000078492 UP000053825 UP000242457 UP000235965 UP000036403 UP000078541 UP000009046 UP000008237 UP000078542 UP000000311 UP000215335 UP000030746 UP000053097 UP000279307 UP000027135 UP000085678 UP000007267 UP000261540 UP000019118 UP000030742 UP000050525 UP000189705 UP000261440 UP000001646 UP000008672 UP000193380 UP000015103 UP000087266 UP000242188 UP000018467 UP000265140 UP000221080 UP000016666 UP000054081 UP000095300 UP000053760 UP000007754 UP000007648 UP000192221 UP000186698 UP000000589 UP000018468 UP000197619 UP000005215 UP000002282 UP000192224 UP000261640
Pfam
Interpro
IPR001611
Leu-rich_rpt
+ More
IPR036859 CAP-Gly_dom_sf
IPR000938 CAP-Gly_domain
IPR032675 LRR_dom_sf
IPR029071 Ubiquitin-like_domsf
IPR027417 P-loop_NTPase
IPR006689 Small_GTPase_ARF/SAR
IPR005225 Small_GTP-bd_dom
IPR004360 Glyas_Fos-R_dOase_dom
IPR029068 Glyas_Bleomycin-R_OHBP_Dase
IPR037523 VOC
IPR004361 Glyoxalase_1
IPR018146 Glyoxalase_1_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR011709 DUF1605
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR007502 Helicase-assoc_dom
IPR000626 Ubiquitin_dom
IPR003591 Leu-rich_rpt_typical-subtyp
IPR021660 DUF3253
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR036859 CAP-Gly_dom_sf
IPR000938 CAP-Gly_domain
IPR032675 LRR_dom_sf
IPR029071 Ubiquitin-like_domsf
IPR027417 P-loop_NTPase
IPR006689 Small_GTPase_ARF/SAR
IPR005225 Small_GTP-bd_dom
IPR004360 Glyas_Fos-R_dOase_dom
IPR029068 Glyas_Bleomycin-R_OHBP_Dase
IPR037523 VOC
IPR004361 Glyoxalase_1
IPR018146 Glyoxalase_1_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR011709 DUF1605
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR007502 Helicase-assoc_dom
IPR000626 Ubiquitin_dom
IPR003591 Leu-rich_rpt_typical-subtyp
IPR021660 DUF3253
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9ISC2
A0A2H1WCF3
A0A2A4JYI3
A0A0N1I4V6
A0A2W1BFL5
A0A212FJE5
+ More
A0A212EWZ1 A0A194RJW2 A0A158N911 D6WI91 A0A1Y1L5Y1 A0A1Y1L4R6 A0A195BUY8 K7IS98 A0A2C9LD34 A0A154P8U1 A0A0N0BFS8 A0A151WNS7 A0A088ALP0 A0A1W4X421 A0A151J375 A0A0L7QRM2 A0A2A3ETV0 A0A2J7QV38 A0A0J7L128 A0A151K114 E0VEM4 E2BBM1 A0A151IBY3 E2AAG3 A0A232EUB5 A0A310SAK1 A0A1B6EEL5 A0A2J7QV34 A0A0P4XVY7 A0A0P4YSW9 A0A0P6IKA5 V4AGM2 A0A026W415 A0A0P5ZMU4 A0A023F7S1 A0A067RA53 A0A0P6AAN6 A0A1S3IDI9 K7G0W2 A0A1S3ICD7 A0A3B3SA87 A0A0P6DXF3 N6TA25 A0A151M2W4 A0A1B6I484 A0A224XMS2 A0A2D4HA23 A0A0A9Y8G9 A0A0P5CE79 A0A2D4HAA5 A0A0P4VTZ4 A0A1U7RAE4 A0A3B4DXG5 R4GAP8 M3XIN9 A0A060Y2L9 A0A1W7RC53 R4G3X9 V9KB66 A0A1S3L6Y8 R0LCY4 A0A210Q0C9 W5LPV5 T1J621 A0A3P8YIC3 J3S5D9 T1D7I4 A0A0F7Z1T7 W5UCG8 U3IDV7 A0A093RIJ3 A0A1I8NRU6 A0A091G566 H0ZJ32 G3WPP0 A0A1W4VMM1 A0A0P5YR06 A0A1W4VMD9 A0A1L8G8N3 Q5U508 Q8CIV8 W5NG15 A0A091Q4U8 A0A0V0G618 A0A218VAM3 A0A2I0UR94 I3MQ76 B4NYV6 A0A1W4ZSA6 A0A3Q3MKF1 A0A0K8VJ60
A0A212EWZ1 A0A194RJW2 A0A158N911 D6WI91 A0A1Y1L5Y1 A0A1Y1L4R6 A0A195BUY8 K7IS98 A0A2C9LD34 A0A154P8U1 A0A0N0BFS8 A0A151WNS7 A0A088ALP0 A0A1W4X421 A0A151J375 A0A0L7QRM2 A0A2A3ETV0 A0A2J7QV38 A0A0J7L128 A0A151K114 E0VEM4 E2BBM1 A0A151IBY3 E2AAG3 A0A232EUB5 A0A310SAK1 A0A1B6EEL5 A0A2J7QV34 A0A0P4XVY7 A0A0P4YSW9 A0A0P6IKA5 V4AGM2 A0A026W415 A0A0P5ZMU4 A0A023F7S1 A0A067RA53 A0A0P6AAN6 A0A1S3IDI9 K7G0W2 A0A1S3ICD7 A0A3B3SA87 A0A0P6DXF3 N6TA25 A0A151M2W4 A0A1B6I484 A0A224XMS2 A0A2D4HA23 A0A0A9Y8G9 A0A0P5CE79 A0A2D4HAA5 A0A0P4VTZ4 A0A1U7RAE4 A0A3B4DXG5 R4GAP8 M3XIN9 A0A060Y2L9 A0A1W7RC53 R4G3X9 V9KB66 A0A1S3L6Y8 R0LCY4 A0A210Q0C9 W5LPV5 T1J621 A0A3P8YIC3 J3S5D9 T1D7I4 A0A0F7Z1T7 W5UCG8 U3IDV7 A0A093RIJ3 A0A1I8NRU6 A0A091G566 H0ZJ32 G3WPP0 A0A1W4VMM1 A0A0P5YR06 A0A1W4VMD9 A0A1L8G8N3 Q5U508 Q8CIV8 W5NG15 A0A091Q4U8 A0A0V0G618 A0A218VAM3 A0A2I0UR94 I3MQ76 B4NYV6 A0A1W4ZSA6 A0A3Q3MKF1 A0A0K8VJ60
PDB
1LPL
E-value=0.000491064,
Score=104
Ontologies
GO
GO:0005525
GO:0043014
GO:0005737
GO:0007023
GO:0007021
GO:0000226
GO:0004462
GO:0046872
GO:0004771
GO:0004806
GO:0004386
GO:0005524
GO:0009791
GO:0014889
GO:0008344
GO:0048589
GO:0048936
GO:0005856
GO:0007052
GO:0007409
GO:0051082
GO:0006457
GO:0008582
GO:0007274
GO:0046785
GO:0005515
GO:0006468
GO:0004672
GO:0005634
GO:0005507
GO:0004713
GO:0016491
GO:0015930
Topology
Subcellular location
Cytoplasm
Cytoskeleton
Cytoskeleton
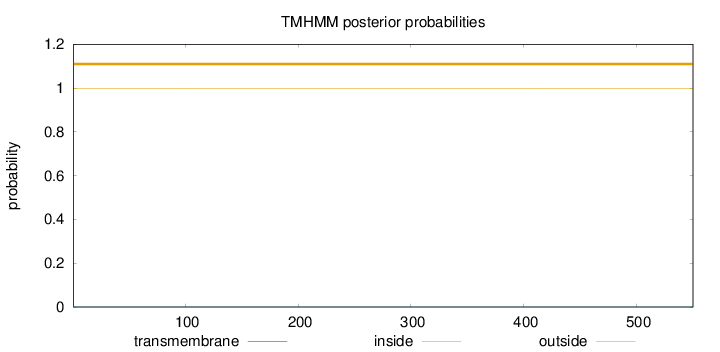
Length:
550
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00025
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00044
outside
1 - 550
Population Genetic Test Statistics
Pi
230.90818
Theta
158.582112
Tajima's D
2.301262
CLR
0.041241
CSRT
0.92320383980801
Interpretation
Uncertain
