Gene
KWMTBOMO14467 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000146
Annotation
PREDICTED:_uncharacterized_protein_LOC101746973_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.21
Sequence
CDS
ATGCTCAGCTCGCAGTCAGCGTCCGAAAGTGAGGCGTCAGAGTCGCGCGCTTCCTCATGGAAACCATGGACAGCTATGGCAGACCTAGCTGAAGCAGTCGACGACCTCATCTGCAGTTTCGACTACATGGACACAGCTATGGACAACCTGGTCATGTTGGTGTTCATATGGATGGTGCTGTCCATAGCGATCATCGCGATCGCCAAATGGGCGTACGGGAGGTTCGCTAAAAAGACGACGGACGCTGATAAACCAAAAATCGATGAGACGAAGAAAACAACGAACGAAATAGTATCCAGTGCTGATAGTGTGCTCGCTACCAGTGCTAAAGTTAAGAGTGCCTCGTTCAAGCCGACTAAAGCGACGGGCTTCGTGCCCGCCACTCCACCGAACAAGAAGAGGTTGGGGCGCATGTCCCCGGGACCAGAGCAGTTGACCGTCAAAAAGCATCAGTCAGTGTCCGTGCCCACATGCACGGGCGGAGATCAGCCGAGCGTGCAATGGGTGAACGATGTACTGACGTGGCTCTACAACGATCTAGTTATCGTAAACGAACTCGTGCAGCAATGGATCTCGTCCATGAACGAGTTCTCGAAGAAATCCGTCGAGGAGCAAGGAATCGGCGTGGAGATAGTACGAGCGTTGGACAGCCACCCGCCCAATATATCGAATGTATTCAGCGAGTGTGACTCCAAGGATGACGTGACGATAACCTGCGACTGTGAAGCCACTCCAGCGTTTCAAGTTAAAGCCTTCAGTCAGCGAGGGGAGAAAGTGGAAGTGTCACATTACAGAGTCAACGTGAATAGGTTCCGCGCCCGCCTCAACGTGGTCTGCGTGACAGATAAGCTCACAGGAGACCTCAAGTTTGACGGGTGGCCAGAAGTCAAAGTGGCACTGGCGCCGGTCGGTCCTTTGAGGCCGAACGCCACTGAATCCAACCTACAGCAGTTGATCAGTGACATTGTCGTGAATGCCTTAAGAAATTCCCATCTGCAGTTTAATCTGTCTCAATACCCGACCTGTCCTAGGCTGAGACGTTACGTTGAGACATCCTCGCGCCTACCCCTGCACTATGATTCCATGTTACAGCACGACCGTCACATGTACACCTCGACTCCGAACTCAACGGCCGGCAGCGGTCTACTCGGAGACAAGAGGTTGCTGGTCAAGATCGTCGGTGCCAAGGACCTGGGAGGTAAAACTGGGTGCATAGCACCGTACTGCGTGGTGGAGATGGACGAGCCTCCCCAGAAGAACCAAACCGGCTTCAAGAAGGAAACCACGAGTCCTCAGTGGGACGAACACTTTTTGTTCGACCTGTCTCCGCAGACCGCGGAGTTGCTCTTTGAGGTCTACGACCGGACCGGACCCCCGTCCCCGGGGGGCGGATCCTCGCAGCACAGGTTCTTAGGTCTGGGCTTGGTCGGTATTGACGAACTGTTGGCGGCGCCCTCCCAGAGACAACAGCTGGCTCTGCAGACCAGGCCTATGGAACAACACGACGTCAGCGGCACCCTCACCGTCGAGTTCCTCTTCATCGACGGCCCCGACATCCCGGACTTCGGCTCCAGACCGGTCAAGATCAAGGAGAGCCTCAGGACCGTGTCCCCTCACGGCCAACTCACCACCACCACCAAGACCACCTTCCATCAGAACGGGATACCAGCACAGGACAACTCTAAGCTGACGGAGTCCGCTCTCAGGGAGCTGGAGCTCAGTCCGACAGGCGCCGCCAACAAGTCCACCCTCATCATACACTCGGTGCAGAGAGAACCCAAGAAGTCTATAATGGTCGAACTAACCGACTCCGGGAAATTCATTGAAGTCGAACGCGCAGACGCCATAGACAAACAGGAAACGGAAGCGAAACCATTAGATAAAATCGAAGAGGCGGTTACAGATAAAAAATCTGTCACCCCGAACCAATCTCCCGGCAAAAGTGTAAGGATCAAAGAGAACGGTAACAAAGTCAACGGAGAGAACAATGATTCGCAAGGAGGCTACGTGAACAACCAGACGGATAACATGTCCCCCGAGCCGGCTCCTAGGAAGAGAAGGAGAGATTTCTTCGGAACTATCAAGAGAAGACTGGGTCGATCTCGTACTCGCGGTCAGTCACTGGAGATGACGGACTCCGAACTGGAGACCCAGAACAGGATGCGTAGCGTCAGTGCCGAGAGGAACTCTCATGAAAAAGCACTGGCAATACCGAACCGCAACGTGTATTCGGCGGGCGCCTCTCCCGCGCAGTCCGGAGACGAATCCAGAACTTCAAGGAGCGAAGTGTCCGGATTCAGTACAGCGTCAGCGAGGACCTTCATTCACGAGGCCTCGACCTTGGTGCTGGAGACTATAGAGGCCGGTATAAAGAAGCATTACCTTGTGCCTCTGGCGATCGCTCAAAGGTCGAAGTGGCGACGGAAGGGCGTCAAACTGCATATTTACAATGAGCACACGTTCATTGCCAAACATCTGAGCGGCGGCACCGTGTGCGCCGTGTGCGACAAGCCGGTGGCACGGCGGCTCGGCAAGCAGGGCTACGAGTGCCGCGACTGCCGGCTGCGCTGCCACAAGCACTGCCACGTGCGTGCGGCCGCCGCCTGCCCCGCCTCCACCGTGCACGACATGGAGCTATCCATGCTCCCCGTGCTAACTGAATAG
Protein
MLSSQSASESEASESRASSWKPWTAMADLAEAVDDLICSFDYMDTAMDNLVMLVFIWMVLSIAIIAIAKWAYGRFAKKTTDADKPKIDETKKTTNEIVSSADSVLATSAKVKSASFKPTKATGFVPATPPNKKRLGRMSPGPEQLTVKKHQSVSVPTCTGGDQPSVQWVNDVLTWLYNDLVIVNELVQQWISSMNEFSKKSVEEQGIGVEIVRALDSHPPNISNVFSECDSKDDVTITCDCEATPAFQVKAFSQRGEKVEVSHYRVNVNRFRARLNVVCVTDKLTGDLKFDGWPEVKVALAPVGPLRPNATESNLQQLISDIVVNALRNSHLQFNLSQYPTCPRLRRYVETSSRLPLHYDSMLQHDRHMYTSTPNSTAGSGLLGDKRLLVKIVGAKDLGGKTGCIAPYCVVEMDEPPQKNQTGFKKETTSPQWDEHFLFDLSPQTAELLFEVYDRTGPPSPGGGSSQHRFLGLGLVGIDELLAAPSQRQQLALQTRPMEQHDVSGTLTVEFLFIDGPDIPDFGSRPVKIKESLRTVSPHGQLTTTTKTTFHQNGIPAQDNSKLTESALRELELSPTGAANKSTLIIHSVQREPKKSIMVELTDSGKFIEVERADAIDKQETEAKPLDKIEEAVTDKKSVTPNQSPGKSVRIKENGNKVNGENNDSQGGYVNNQTDNMSPEPAPRKRRRDFFGTIKRRLGRSRTRGQSLEMTDSELETQNRMRSVSAERNSHEKALAIPNRNVYSAGASPAQSGDESRTSRSEVSGFSTASARTFIHEASTLVLETIEAGIKKHYLVPLAIAQRSKWRRKGVKLHIYNEHTFIAKHLSGGTVCAVCDKPVARRLGKQGYECRDCRLRCHKHCHVRAAAACPASTVHDMELSMLPVLTE
Summary
Uniprot
A0A212F5M5
A0A0N1IPR1
A0A2A4J1Z7
A0A2A4J2C8
A0A2A4J3D0
A0A2A4J1R2
+ More
A0A2A4J3N8 A0A2A4J3J8 A0A1Y1KZI0 A0A1Y1KUL7 A0A1Y1L1X5 D6WH87 A0A1Y1KUN5 A0A1Y1KUT8 A0A1Y1L0G1 A0A023F1L1 A0A0K8SUD9 A0A1B6F688 A0A1B6MKV8 A0A0K8TD59 A0A0L7RCK5 E2C538 A0A087ZSB8 A0A151WZV6 F4WUA2 A0A026X0P2 A0A3L8DST1 A0A1B6IVJ5 A0A195FKE8 A0A232F7S8 A0A195C7G2 A0A1B6JST6 E2AZ18 A0A2A3EN59 A0A1B0CL68 A0A1L8DNB9 A0A1L8DKK9 A0A1L8DNA3 A0A1L8DKI2 K7J3I6 A0A1L8DKJ3 A0A1L8DNC0 A0A1L8DNA7 A0A1L8DKT3 A0A1L8DKW2 A0A1L8DKN8 A0A1L8DKS5 N6TP12 N6STF9 U4TVA9 E9IH50 E0VR85 A0A0T6BHW7 A0A1L8DKG5 A0A1L8DKV3 A0A1L8DKQ0 A0A1L8DKP3 A0A0Q5VV57 A0A0Q5VL02 A0A1I8NUL0 A0A1I8NUJ7 A0A1I8NUK0 A0A0K8TBJ6 A0A1W4VHX3 A0A1L8DKC3 A0A1I8NUK2 A0A0Q5VZW5 A0A0Q5VL88 A0A0Q9X8T5 A0A034VLF5 A0A0Q9XLH0 A0A0R1DXC6 A0A0Q9XM18 A0A0Q5VYQ5 A0A0R1DXF8 A0A1I8NUJ8 A0A0Q9XB87 A0A034VLG7 B3NJI8 B4JVM5 A0A3B0IY92 A0A0Q9XIV8 A0A0Q5VL21 A0A0R1DYJ7
A0A2A4J3N8 A0A2A4J3J8 A0A1Y1KZI0 A0A1Y1KUL7 A0A1Y1L1X5 D6WH87 A0A1Y1KUN5 A0A1Y1KUT8 A0A1Y1L0G1 A0A023F1L1 A0A0K8SUD9 A0A1B6F688 A0A1B6MKV8 A0A0K8TD59 A0A0L7RCK5 E2C538 A0A087ZSB8 A0A151WZV6 F4WUA2 A0A026X0P2 A0A3L8DST1 A0A1B6IVJ5 A0A195FKE8 A0A232F7S8 A0A195C7G2 A0A1B6JST6 E2AZ18 A0A2A3EN59 A0A1B0CL68 A0A1L8DNB9 A0A1L8DKK9 A0A1L8DNA3 A0A1L8DKI2 K7J3I6 A0A1L8DKJ3 A0A1L8DNC0 A0A1L8DNA7 A0A1L8DKT3 A0A1L8DKW2 A0A1L8DKN8 A0A1L8DKS5 N6TP12 N6STF9 U4TVA9 E9IH50 E0VR85 A0A0T6BHW7 A0A1L8DKG5 A0A1L8DKV3 A0A1L8DKQ0 A0A1L8DKP3 A0A0Q5VV57 A0A0Q5VL02 A0A1I8NUL0 A0A1I8NUJ7 A0A1I8NUK0 A0A0K8TBJ6 A0A1W4VHX3 A0A1L8DKC3 A0A1I8NUK2 A0A0Q5VZW5 A0A0Q5VL88 A0A0Q9X8T5 A0A034VLF5 A0A0Q9XLH0 A0A0R1DXC6 A0A0Q9XM18 A0A0Q5VYQ5 A0A0R1DXF8 A0A1I8NUJ8 A0A0Q9XB87 A0A034VLG7 B3NJI8 B4JVM5 A0A3B0IY92 A0A0Q9XIV8 A0A0Q5VL21 A0A0R1DYJ7
Pubmed
EMBL
AGBW02010170
OWR49037.1
KQ460128
KPJ17572.1
NWSH01003634
PCG66021.1
+ More
PCG66019.1 PCG66024.1 PCG66015.1 PCG66022.1 PCG66020.1 GEZM01073433 JAV65095.1 GEZM01073435 JAV65093.1 GEZM01073431 JAV65097.1 KQ971330 EEZ99736.1 GEZM01073436 JAV65092.1 GEZM01073434 JAV65094.1 GEZM01073432 JAV65096.1 GBBI01003524 JAC15188.1 GBRD01008893 JAG56928.1 GECZ01024406 GECZ01010708 JAS45363.1 JAS59061.1 GEBQ01003454 JAT36523.1 GBRD01002362 JAG63459.1 KQ414615 KOC68722.1 GL452737 EFN76946.1 KQ982628 KYQ53442.1 GL888355 EGI62210.1 KK107054 EZA61618.1 QOIP01000004 RLU23464.1 GECU01016751 JAS90955.1 KQ981522 KYN40737.1 NNAY01000788 OXU26508.1 KQ978143 KYM96814.1 GECU01005479 JAT02228.1 GL444070 EFN61313.1 KZ288204 PBC33168.1 AJWK01016976 AJWK01016977 AJWK01016978 AJWK01016979 AJWK01016980 GFDF01006135 JAV07949.1 GFDF01007102 JAV06982.1 GFDF01006131 JAV07953.1 GFDF01007101 JAV06983.1 GFDF01007108 JAV06976.1 GFDF01006132 JAV07952.1 GFDF01006136 JAV07948.1 GFDF01007107 JAV06977.1 GFDF01007104 JAV06980.1 GFDF01007103 JAV06981.1 GFDF01007109 JAV06975.1 APGK01057542 APGK01057543 KB741280 ENN70970.1 ENN70969.1 KB631620 ERL84737.1 GL763174 EFZ20103.1 DS235459 EEB15891.1 LJIG01000019 KRT86921.1 GFDF01007138 JAV06946.1 GFDF01007110 JAV06974.1 GFDF01007137 JAV06947.1 GFDF01007147 JAV06937.1 CH954179 KQS62139.1 KQS62134.1 GBRD01002898 GBRD01002363 JAG62923.1 GFDF01007175 JAV06909.1 KQS62143.1 KQS62129.1 CH933808 KRG04822.1 GAKP01015668 JAC43284.1 KRG04824.1 CM000158 KRJ99920.1 KRG04835.1 KQS62146.1 KRJ99929.1 KRG04825.1 GAKP01015653 JAC43299.1 EDV55178.1 CH916375 EDV98493.1 OUUW01000001 SPP72687.1 KRG04830.1 KQS62150.1 KRJ99926.1
PCG66019.1 PCG66024.1 PCG66015.1 PCG66022.1 PCG66020.1 GEZM01073433 JAV65095.1 GEZM01073435 JAV65093.1 GEZM01073431 JAV65097.1 KQ971330 EEZ99736.1 GEZM01073436 JAV65092.1 GEZM01073434 JAV65094.1 GEZM01073432 JAV65096.1 GBBI01003524 JAC15188.1 GBRD01008893 JAG56928.1 GECZ01024406 GECZ01010708 JAS45363.1 JAS59061.1 GEBQ01003454 JAT36523.1 GBRD01002362 JAG63459.1 KQ414615 KOC68722.1 GL452737 EFN76946.1 KQ982628 KYQ53442.1 GL888355 EGI62210.1 KK107054 EZA61618.1 QOIP01000004 RLU23464.1 GECU01016751 JAS90955.1 KQ981522 KYN40737.1 NNAY01000788 OXU26508.1 KQ978143 KYM96814.1 GECU01005479 JAT02228.1 GL444070 EFN61313.1 KZ288204 PBC33168.1 AJWK01016976 AJWK01016977 AJWK01016978 AJWK01016979 AJWK01016980 GFDF01006135 JAV07949.1 GFDF01007102 JAV06982.1 GFDF01006131 JAV07953.1 GFDF01007101 JAV06983.1 GFDF01007108 JAV06976.1 GFDF01006132 JAV07952.1 GFDF01006136 JAV07948.1 GFDF01007107 JAV06977.1 GFDF01007104 JAV06980.1 GFDF01007103 JAV06981.1 GFDF01007109 JAV06975.1 APGK01057542 APGK01057543 KB741280 ENN70970.1 ENN70969.1 KB631620 ERL84737.1 GL763174 EFZ20103.1 DS235459 EEB15891.1 LJIG01000019 KRT86921.1 GFDF01007138 JAV06946.1 GFDF01007110 JAV06974.1 GFDF01007137 JAV06947.1 GFDF01007147 JAV06937.1 CH954179 KQS62139.1 KQS62134.1 GBRD01002898 GBRD01002363 JAG62923.1 GFDF01007175 JAV06909.1 KQS62143.1 KQS62129.1 CH933808 KRG04822.1 GAKP01015668 JAC43284.1 KRG04824.1 CM000158 KRJ99920.1 KRG04835.1 KQS62146.1 KRJ99929.1 KRG04825.1 GAKP01015653 JAC43299.1 EDV55178.1 CH916375 EDV98493.1 OUUW01000001 SPP72687.1 KRG04830.1 KQS62150.1 KRJ99926.1
Proteomes
UP000007151
UP000053240
UP000218220
UP000007266
UP000053825
UP000008237
+ More
UP000005203 UP000075809 UP000007755 UP000053097 UP000279307 UP000078541 UP000215335 UP000078542 UP000000311 UP000242457 UP000092461 UP000002358 UP000019118 UP000030742 UP000009046 UP000008711 UP000095300 UP000192221 UP000009192 UP000002282 UP000001070 UP000268350
UP000005203 UP000075809 UP000007755 UP000053097 UP000279307 UP000078541 UP000215335 UP000078542 UP000000311 UP000242457 UP000092461 UP000002358 UP000019118 UP000030742 UP000009046 UP000008711 UP000095300 UP000192221 UP000009192 UP000002282 UP000001070 UP000268350
Gene 3D
CDD
ProteinModelPortal
A0A212F5M5
A0A0N1IPR1
A0A2A4J1Z7
A0A2A4J2C8
A0A2A4J3D0
A0A2A4J1R2
+ More
A0A2A4J3N8 A0A2A4J3J8 A0A1Y1KZI0 A0A1Y1KUL7 A0A1Y1L1X5 D6WH87 A0A1Y1KUN5 A0A1Y1KUT8 A0A1Y1L0G1 A0A023F1L1 A0A0K8SUD9 A0A1B6F688 A0A1B6MKV8 A0A0K8TD59 A0A0L7RCK5 E2C538 A0A087ZSB8 A0A151WZV6 F4WUA2 A0A026X0P2 A0A3L8DST1 A0A1B6IVJ5 A0A195FKE8 A0A232F7S8 A0A195C7G2 A0A1B6JST6 E2AZ18 A0A2A3EN59 A0A1B0CL68 A0A1L8DNB9 A0A1L8DKK9 A0A1L8DNA3 A0A1L8DKI2 K7J3I6 A0A1L8DKJ3 A0A1L8DNC0 A0A1L8DNA7 A0A1L8DKT3 A0A1L8DKW2 A0A1L8DKN8 A0A1L8DKS5 N6TP12 N6STF9 U4TVA9 E9IH50 E0VR85 A0A0T6BHW7 A0A1L8DKG5 A0A1L8DKV3 A0A1L8DKQ0 A0A1L8DKP3 A0A0Q5VV57 A0A0Q5VL02 A0A1I8NUL0 A0A1I8NUJ7 A0A1I8NUK0 A0A0K8TBJ6 A0A1W4VHX3 A0A1L8DKC3 A0A1I8NUK2 A0A0Q5VZW5 A0A0Q5VL88 A0A0Q9X8T5 A0A034VLF5 A0A0Q9XLH0 A0A0R1DXC6 A0A0Q9XM18 A0A0Q5VYQ5 A0A0R1DXF8 A0A1I8NUJ8 A0A0Q9XB87 A0A034VLG7 B3NJI8 B4JVM5 A0A3B0IY92 A0A0Q9XIV8 A0A0Q5VL21 A0A0R1DYJ7
A0A2A4J3N8 A0A2A4J3J8 A0A1Y1KZI0 A0A1Y1KUL7 A0A1Y1L1X5 D6WH87 A0A1Y1KUN5 A0A1Y1KUT8 A0A1Y1L0G1 A0A023F1L1 A0A0K8SUD9 A0A1B6F688 A0A1B6MKV8 A0A0K8TD59 A0A0L7RCK5 E2C538 A0A087ZSB8 A0A151WZV6 F4WUA2 A0A026X0P2 A0A3L8DST1 A0A1B6IVJ5 A0A195FKE8 A0A232F7S8 A0A195C7G2 A0A1B6JST6 E2AZ18 A0A2A3EN59 A0A1B0CL68 A0A1L8DNB9 A0A1L8DKK9 A0A1L8DNA3 A0A1L8DKI2 K7J3I6 A0A1L8DKJ3 A0A1L8DNC0 A0A1L8DNA7 A0A1L8DKT3 A0A1L8DKW2 A0A1L8DKN8 A0A1L8DKS5 N6TP12 N6STF9 U4TVA9 E9IH50 E0VR85 A0A0T6BHW7 A0A1L8DKG5 A0A1L8DKV3 A0A1L8DKQ0 A0A1L8DKP3 A0A0Q5VV57 A0A0Q5VL02 A0A1I8NUL0 A0A1I8NUJ7 A0A1I8NUK0 A0A0K8TBJ6 A0A1W4VHX3 A0A1L8DKC3 A0A1I8NUK2 A0A0Q5VZW5 A0A0Q5VL88 A0A0Q9X8T5 A0A034VLF5 A0A0Q9XLH0 A0A0R1DXC6 A0A0Q9XM18 A0A0Q5VYQ5 A0A0R1DXF8 A0A1I8NUJ8 A0A0Q9XB87 A0A034VLG7 B3NJI8 B4JVM5 A0A3B0IY92 A0A0Q9XIV8 A0A0Q5VL21 A0A0R1DYJ7
PDB
2YUU
E-value=3.21771e-05,
Score=116
Ontologies
GO
PANTHER
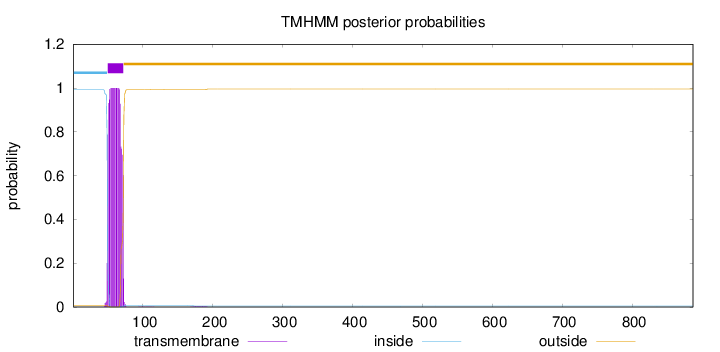
Topology
Length:
887
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.8278499999999
Exp number, first 60 AAs:
10.9225
Total prob of N-in:
0.99192
POSSIBLE N-term signal
sequence
inside
1 - 49
TMhelix
50 - 72
outside
73 - 887
Population Genetic Test Statistics
Pi
221.330997
Theta
162.66938
Tajima's D
1.36533
CLR
0.294338
CSRT
0.75961201939903
Interpretation
Uncertain
