Pre Gene Modal
BGIBMGA000081
Annotation
DNA_polymerase_epsilon_subunit_3_[Bombyx_mandarina]
Full name
DNA polymerase epsilon subunit 3
Alternative Name
DNA polymerase II subunit 3
DNA polymerase epsilon subunit p17
Arsenic-transactivated protein
Chromatin accessibility complex 17 kDa protein
NF-YB-like protein
YB-like protein 1
DNA polymerase epsilon subunit p17
Arsenic-transactivated protein
Chromatin accessibility complex 17 kDa protein
NF-YB-like protein
YB-like protein 1
Location in the cell
Cytoplasmic Reliability : 1.109 Nuclear Reliability : 1.21
Sequence
CDS
ATGGCTGAAAAACTCGAAGATTTGAATTTACCACTCACAGTTGTAACTAGAATCGTAAAAGAAGCTCTTCCCAGTGGCGTATCGATATCAAAAGAGGCGCGAACTGGCCTCGCTAAGGCGGCTTCAGTTTTTGTACTTTATGTGACTTCAGCAGCTACAAATATTGTTAAGAACAAAAAAAGGAAGGCTCTCACTGGGCAGGACGTGATAGATGCTATGAAAGACATCGAATTTGATAGATTTGTGGAACCTTTAGGAGAAGCTCTGGAACATTACAAGACCATGGCTTCGGCACGAAAATCCGCTTCTATGAAGAAAAAAGATGAACAAGAAGACGCTGAGATTATAGAAGACGATTGA
Protein
MAEKLEDLNLPLTVVTRIVKEALPSGVSISKEARTGLAKAASVFVLYVTSAATNIVKNKKRKALTGQDVIDAMKDIEFDRFVEPLGEALEHYKTMASARKSASMKKKDEQEDAEIIEDD
Summary
Description
Forms a complex with DNA polymerase epsilon subunit CHRAC1 and binds naked DNA, which is then incorporated into chromatin, aided by the nucleosome-remodeling activity of ISWI/SNF2H and ACF1.
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Subunit
Component of the epsilon DNA polymerase complex consisting of four subunits: POLE, POLE2, POLE3 and POLE4. Interaction with POLE4 is a prerequisite for further binding with POLE and POLE2. Interacts with CHRAC1. Together with CHRAC1, ACF1 and ISWI/SNF2H proteins, it forms the ISWI chromatin-remodeling complex, CHRAC (By similarity).
Component of the epsilon DNA polymerase complex consisting of four subunits: POLE, POLE2, POLE3 and POLE4. Interaction with POLE4 is a prerequisite for further binding with POLE and POLE2. Interacts with CHRAC1. Together with CHRAC1, ACF1 and ISWI/SNF2H proteins, it forms the ISWI chromatin-remodeling complex, CHRAC.
Component of the epsilon DNA polymerase complex consisting of four subunits: POLE, POLE2, POLE3 and POLE4. Interaction with POLE4 is a prerequisite for further binding with POLE and POLE2. Interacts with CHRAC1. Together with CHRAC1, ACF1 and ISWI/SNF2H proteins, it forms the ISWI chromatin-remodeling complex, CHRAC.
Keywords
Acetylation
Coiled coil
Complete proteome
DNA-binding
DNA-directed DNA polymerase
Nucleotidyltransferase
Nucleus
Phosphoprotein
Reference proteome
Transferase
Direct protein sequencing
Polymorphism
Feature
chain DNA polymerase epsilon subunit 3
sequence variant In dbSNP:rs36023979.
sequence variant In dbSNP:rs36023979.
Uniprot
A0A075DA27
A0A075D847
H9IS55
A0A075DA16
A0A075DB57
A0A075D852
+ More
A0A075DA32 A0A075DB67 A0A075D9D4 A0A2H1VXU8 A0A2W1BC06 A0A2A4K179 A0A212F3Z4 A0A0L7LAB6 S4P4D0 A0A0N1I9H0 A0A0N0P9M8 E2BSU6 A0A2A3E3D6 A0A088A5N7 A0A0L7R9C2 A0A154PMQ9 A0A0J7KXT9 E2AN99 A0A0N0BEN9 A0A310SH20 A0A151WQV3 F4W555 A0A151I4Q7 A0A158NYZ2 A0A195FT22 T1JJN7 A0A195D0P5 A0A151JLM6 A0A341CVR8 A0A2U3V899 A0A340WQ97 A0A2Y9EVK6 A0A2Y9NP82 E9J7J3 A0A384BG50 A0A1U7QM76 A0A091D6T8 A0A1S3KCV7 L5LG49 K9IG13 L9L6T9 A0A3L7I795 F1SN88 L8I4S1 G1PS53 Q642A5 A0A2K6DZM7 A0A2K6MTL7 G3QRK2 G1S438 A0A2K5YRF9 A0A0D9RJ77 A0A2K5DGQ8 A0A2K6Q9X0 A0A2K5NWD2 H0XNA5 W5P8E2 A0A2K5RX40 A0A2K5HSW4 A0A212CN04 A0A2K6UY63 A0A2J8WRX2 G7PRM1 G2HEE7 F7EVY7 A0A024R829 Q5R4W3 Q9NRF9 Q3SZN5 A0A384DGV8 A0A2Y9HCI1 A0A2U3XUN3 A0A2Y9J7A5 M3Y960 A0A3Q7W934 D2H2I7 A0A2Y9DDQ0 A0A2R9CDA1 M3XC08 A0A2K6GSW9 R7TVV9 A0A3Q2ICV7 A0A3Q7QQ23 A0A2U3VPQ6 A0A0R4J091 A0A164NZC7 A0A286XAE1 L5KA46 A0A1S2ZYZ9 J9P0Z6 A0A1U7TP20 Q9JKP7 G5B1Z8 F6TIZ6
A0A075DA32 A0A075DB67 A0A075D9D4 A0A2H1VXU8 A0A2W1BC06 A0A2A4K179 A0A212F3Z4 A0A0L7LAB6 S4P4D0 A0A0N1I9H0 A0A0N0P9M8 E2BSU6 A0A2A3E3D6 A0A088A5N7 A0A0L7R9C2 A0A154PMQ9 A0A0J7KXT9 E2AN99 A0A0N0BEN9 A0A310SH20 A0A151WQV3 F4W555 A0A151I4Q7 A0A158NYZ2 A0A195FT22 T1JJN7 A0A195D0P5 A0A151JLM6 A0A341CVR8 A0A2U3V899 A0A340WQ97 A0A2Y9EVK6 A0A2Y9NP82 E9J7J3 A0A384BG50 A0A1U7QM76 A0A091D6T8 A0A1S3KCV7 L5LG49 K9IG13 L9L6T9 A0A3L7I795 F1SN88 L8I4S1 G1PS53 Q642A5 A0A2K6DZM7 A0A2K6MTL7 G3QRK2 G1S438 A0A2K5YRF9 A0A0D9RJ77 A0A2K5DGQ8 A0A2K6Q9X0 A0A2K5NWD2 H0XNA5 W5P8E2 A0A2K5RX40 A0A2K5HSW4 A0A212CN04 A0A2K6UY63 A0A2J8WRX2 G7PRM1 G2HEE7 F7EVY7 A0A024R829 Q5R4W3 Q9NRF9 Q3SZN5 A0A384DGV8 A0A2Y9HCI1 A0A2U3XUN3 A0A2Y9J7A5 M3Y960 A0A3Q7W934 D2H2I7 A0A2Y9DDQ0 A0A2R9CDA1 M3XC08 A0A2K6GSW9 R7TVV9 A0A3Q2ICV7 A0A3Q7QQ23 A0A2U3VPQ6 A0A0R4J091 A0A164NZC7 A0A286XAE1 L5KA46 A0A1S2ZYZ9 J9P0Z6 A0A1U7TP20 Q9JKP7 G5B1Z8 F6TIZ6
EC Number
2.7.7.7
Pubmed
25123546
19121390
28756777
22118469
26227816
23622113
+ More
26354079 20798317 21719571 21347285 21282665 23385571 29704459 22751099 21993624 15489334 22673903 22398555 25362486 20809919 17194215 22002653 16136131 21484476 17431167 25319552 11181995 10880450 10801849 14702039 15164053 18669648 19413330 20068231 21269460 21406692 16305752 24813606 20010809 22722832 17975172 23254933 19892987 12040188 19468303 21183079 23258410 16341006 11000277 21993625 25243066
26354079 20798317 21719571 21347285 21282665 23385571 29704459 22751099 21993624 15489334 22673903 22398555 25362486 20809919 17194215 22002653 16136131 21484476 17431167 25319552 11181995 10880450 10801849 14702039 15164053 18669648 19413330 20068231 21269460 21406692 16305752 24813606 20010809 22722832 17975172 23254933 19892987 12040188 19468303 21183079 23258410 16341006 11000277 21993625 25243066
EMBL
KF703558
KF703559
KF703560
KF703561
AHH91062.1
AHH91063.1
+ More
AHH91064.1 AHH91065.1 KF703557 AHH91061.1 BABH01024255 KF703567 KF703568 KF703569 KF703570 AHH91071.1 AHH91072.1 AHH91073.1 AHH91074.1 KF703565 AHH91069.1 KF703556 AHH91060.1 KF703562 AHH91066.1 KF703563 AHH91067.1 KF703566 AHH91070.1 KF703564 AHH91068.1 ODYU01005114 SOQ45660.1 KZ150181 PZC72508.1 NWSH01000293 PCG77678.1 AGBW02010484 OWR48433.1 JTDY01002005 KOB72350.1 GAIX01009102 JAA83458.1 KQ460128 KPJ17571.1 KQ459286 KPJ02017.1 GL450269 EFN81238.1 KZ288400 PBC26263.1 KQ414627 KOC67472.1 KQ434972 KZC12744.1 LBMM01002238 KMQ95088.1 GL441149 EFN65101.1 KQ435828 KOX71891.1 KQ769940 OAD52732.1 KQ982821 KYQ50188.1 GL887605 EGI70651.1 KQ976448 KYM85876.1 ADTU01004412 KQ981276 KYN43591.1 JH431939 KQ977004 KYN06488.1 KQ978993 KYN26690.1 GL768539 EFZ11212.1 KN123095 KFO26762.1 KB112777 ELK24608.1 GABZ01008375 JAA45150.1 KB320489 ELW70603.1 RAZU01000090 RLQ74056.1 AEMK02000005 JH881972 ELR51510.1 AAPE02016217 BC081988 BC083800 CABD030065952 ADFV01019711 AQIB01022397 AAQR03034664 AMGL01047119 MKHE01000016 OWK07361.1 NDHI03003380 PNJ72484.1 PNJ72485.1 AQIA01023729 AB170211 CM001290 BAE87274.1 EHH57137.1 AACZ04069042 AK305111 GABC01006272 GABF01008439 GABD01000857 GABE01000340 NBAG03000232 BAK62105.1 JAA05066.1 JAA13706.1 JAA32243.1 JAA44399.1 PNI69606.1 PNI69607.1 JSUE03014414 JU320645 JU320646 JU470579 JU470580 JV043795 JV043796 CM001267 AFE64401.1 AFH27383.1 AFI33866.1 EHH23849.1 CH471090 EAW87382.1 CR861128 AF226077 AF261689 AY720898 AK074629 AK074762 AK074782 DQ072116 AL137066 BC003166 BC004170 BT030577 BC102772 AEYP01022990 ACTA01178982 GL192443 EFB15511.1 AJFE02119091 AANG04004382 AMQN01010745 KB308469 ELT97819.1 AL732594 CH466527 EDL31135.1 LRGB01002761 KZS06376.1 AAKN02050013 KB030911 ELK08242.1 AAEX03008085 AF230806 BC024996 JH168050 GEBF01001156 EHB03300.1 JAO02477.1 GAMT01000156 GAMS01004215 GAMS01004214 GAMR01006129 GAMR01006128 GAMQ01004390 JAB11705.1 JAB18921.1 JAB27803.1 JAB37461.1
AHH91064.1 AHH91065.1 KF703557 AHH91061.1 BABH01024255 KF703567 KF703568 KF703569 KF703570 AHH91071.1 AHH91072.1 AHH91073.1 AHH91074.1 KF703565 AHH91069.1 KF703556 AHH91060.1 KF703562 AHH91066.1 KF703563 AHH91067.1 KF703566 AHH91070.1 KF703564 AHH91068.1 ODYU01005114 SOQ45660.1 KZ150181 PZC72508.1 NWSH01000293 PCG77678.1 AGBW02010484 OWR48433.1 JTDY01002005 KOB72350.1 GAIX01009102 JAA83458.1 KQ460128 KPJ17571.1 KQ459286 KPJ02017.1 GL450269 EFN81238.1 KZ288400 PBC26263.1 KQ414627 KOC67472.1 KQ434972 KZC12744.1 LBMM01002238 KMQ95088.1 GL441149 EFN65101.1 KQ435828 KOX71891.1 KQ769940 OAD52732.1 KQ982821 KYQ50188.1 GL887605 EGI70651.1 KQ976448 KYM85876.1 ADTU01004412 KQ981276 KYN43591.1 JH431939 KQ977004 KYN06488.1 KQ978993 KYN26690.1 GL768539 EFZ11212.1 KN123095 KFO26762.1 KB112777 ELK24608.1 GABZ01008375 JAA45150.1 KB320489 ELW70603.1 RAZU01000090 RLQ74056.1 AEMK02000005 JH881972 ELR51510.1 AAPE02016217 BC081988 BC083800 CABD030065952 ADFV01019711 AQIB01022397 AAQR03034664 AMGL01047119 MKHE01000016 OWK07361.1 NDHI03003380 PNJ72484.1 PNJ72485.1 AQIA01023729 AB170211 CM001290 BAE87274.1 EHH57137.1 AACZ04069042 AK305111 GABC01006272 GABF01008439 GABD01000857 GABE01000340 NBAG03000232 BAK62105.1 JAA05066.1 JAA13706.1 JAA32243.1 JAA44399.1 PNI69606.1 PNI69607.1 JSUE03014414 JU320645 JU320646 JU470579 JU470580 JV043795 JV043796 CM001267 AFE64401.1 AFH27383.1 AFI33866.1 EHH23849.1 CH471090 EAW87382.1 CR861128 AF226077 AF261689 AY720898 AK074629 AK074762 AK074782 DQ072116 AL137066 BC003166 BC004170 BT030577 BC102772 AEYP01022990 ACTA01178982 GL192443 EFB15511.1 AJFE02119091 AANG04004382 AMQN01010745 KB308469 ELT97819.1 AL732594 CH466527 EDL31135.1 LRGB01002761 KZS06376.1 AAKN02050013 KB030911 ELK08242.1 AAEX03008085 AF230806 BC024996 JH168050 GEBF01001156 EHB03300.1 JAO02477.1 GAMT01000156 GAMS01004215 GAMS01004214 GAMR01006129 GAMR01006128 GAMQ01004390 JAB11705.1 JAB18921.1 JAB27803.1 JAB37461.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
+ More
UP000008237 UP000242457 UP000005203 UP000053825 UP000076502 UP000036403 UP000000311 UP000053105 UP000075809 UP000007755 UP000078540 UP000005205 UP000078541 UP000078542 UP000078492 UP000252040 UP000245320 UP000265300 UP000248484 UP000248483 UP000261681 UP000189706 UP000028990 UP000085678 UP000011518 UP000273346 UP000008227 UP000001074 UP000002494 UP000233120 UP000233180 UP000001519 UP000001073 UP000233140 UP000029965 UP000233020 UP000233200 UP000233060 UP000005225 UP000002356 UP000233040 UP000233080 UP000233220 UP000009130 UP000233100 UP000002277 UP000006718 UP000001595 UP000005640 UP000009136 UP000261680 UP000291021 UP000248481 UP000245341 UP000248482 UP000000715 UP000286642 UP000008912 UP000248480 UP000240080 UP000011712 UP000233160 UP000014760 UP000002281 UP000286641 UP000245340 UP000000589 UP000076858 UP000005447 UP000010552 UP000079721 UP000002254 UP000189704 UP000006813 UP000008225
UP000008237 UP000242457 UP000005203 UP000053825 UP000076502 UP000036403 UP000000311 UP000053105 UP000075809 UP000007755 UP000078540 UP000005205 UP000078541 UP000078542 UP000078492 UP000252040 UP000245320 UP000265300 UP000248484 UP000248483 UP000261681 UP000189706 UP000028990 UP000085678 UP000011518 UP000273346 UP000008227 UP000001074 UP000002494 UP000233120 UP000233180 UP000001519 UP000001073 UP000233140 UP000029965 UP000233020 UP000233200 UP000233060 UP000005225 UP000002356 UP000233040 UP000233080 UP000233220 UP000009130 UP000233100 UP000002277 UP000006718 UP000001595 UP000005640 UP000009136 UP000261680 UP000291021 UP000248481 UP000245341 UP000248482 UP000000715 UP000286642 UP000008912 UP000248480 UP000240080 UP000011712 UP000233160 UP000014760 UP000002281 UP000286641 UP000245340 UP000000589 UP000076858 UP000005447 UP000010552 UP000079721 UP000002254 UP000189704 UP000006813 UP000008225
Pfam
PF00808 CBFD_NFYB_HMF
SUPFAM
SSF47113
SSF47113
Gene 3D
ProteinModelPortal
A0A075DA27
A0A075D847
H9IS55
A0A075DA16
A0A075DB57
A0A075D852
+ More
A0A075DA32 A0A075DB67 A0A075D9D4 A0A2H1VXU8 A0A2W1BC06 A0A2A4K179 A0A212F3Z4 A0A0L7LAB6 S4P4D0 A0A0N1I9H0 A0A0N0P9M8 E2BSU6 A0A2A3E3D6 A0A088A5N7 A0A0L7R9C2 A0A154PMQ9 A0A0J7KXT9 E2AN99 A0A0N0BEN9 A0A310SH20 A0A151WQV3 F4W555 A0A151I4Q7 A0A158NYZ2 A0A195FT22 T1JJN7 A0A195D0P5 A0A151JLM6 A0A341CVR8 A0A2U3V899 A0A340WQ97 A0A2Y9EVK6 A0A2Y9NP82 E9J7J3 A0A384BG50 A0A1U7QM76 A0A091D6T8 A0A1S3KCV7 L5LG49 K9IG13 L9L6T9 A0A3L7I795 F1SN88 L8I4S1 G1PS53 Q642A5 A0A2K6DZM7 A0A2K6MTL7 G3QRK2 G1S438 A0A2K5YRF9 A0A0D9RJ77 A0A2K5DGQ8 A0A2K6Q9X0 A0A2K5NWD2 H0XNA5 W5P8E2 A0A2K5RX40 A0A2K5HSW4 A0A212CN04 A0A2K6UY63 A0A2J8WRX2 G7PRM1 G2HEE7 F7EVY7 A0A024R829 Q5R4W3 Q9NRF9 Q3SZN5 A0A384DGV8 A0A2Y9HCI1 A0A2U3XUN3 A0A2Y9J7A5 M3Y960 A0A3Q7W934 D2H2I7 A0A2Y9DDQ0 A0A2R9CDA1 M3XC08 A0A2K6GSW9 R7TVV9 A0A3Q2ICV7 A0A3Q7QQ23 A0A2U3VPQ6 A0A0R4J091 A0A164NZC7 A0A286XAE1 L5KA46 A0A1S2ZYZ9 J9P0Z6 A0A1U7TP20 Q9JKP7 G5B1Z8 F6TIZ6
A0A075DA32 A0A075DB67 A0A075D9D4 A0A2H1VXU8 A0A2W1BC06 A0A2A4K179 A0A212F3Z4 A0A0L7LAB6 S4P4D0 A0A0N1I9H0 A0A0N0P9M8 E2BSU6 A0A2A3E3D6 A0A088A5N7 A0A0L7R9C2 A0A154PMQ9 A0A0J7KXT9 E2AN99 A0A0N0BEN9 A0A310SH20 A0A151WQV3 F4W555 A0A151I4Q7 A0A158NYZ2 A0A195FT22 T1JJN7 A0A195D0P5 A0A151JLM6 A0A341CVR8 A0A2U3V899 A0A340WQ97 A0A2Y9EVK6 A0A2Y9NP82 E9J7J3 A0A384BG50 A0A1U7QM76 A0A091D6T8 A0A1S3KCV7 L5LG49 K9IG13 L9L6T9 A0A3L7I795 F1SN88 L8I4S1 G1PS53 Q642A5 A0A2K6DZM7 A0A2K6MTL7 G3QRK2 G1S438 A0A2K5YRF9 A0A0D9RJ77 A0A2K5DGQ8 A0A2K6Q9X0 A0A2K5NWD2 H0XNA5 W5P8E2 A0A2K5RX40 A0A2K5HSW4 A0A212CN04 A0A2K6UY63 A0A2J8WRX2 G7PRM1 G2HEE7 F7EVY7 A0A024R829 Q5R4W3 Q9NRF9 Q3SZN5 A0A384DGV8 A0A2Y9HCI1 A0A2U3XUN3 A0A2Y9J7A5 M3Y960 A0A3Q7W934 D2H2I7 A0A2Y9DDQ0 A0A2R9CDA1 M3XC08 A0A2K6GSW9 R7TVV9 A0A3Q2ICV7 A0A3Q7QQ23 A0A2U3VPQ6 A0A0R4J091 A0A164NZC7 A0A286XAE1 L5KA46 A0A1S2ZYZ9 J9P0Z6 A0A1U7TP20 Q9JKP7 G5B1Z8 F6TIZ6
PDB
2BYM
E-value=1.51465e-13,
Score=178
Ontologies
PATHWAY
GO
Topology
Subcellular location
Nucleus
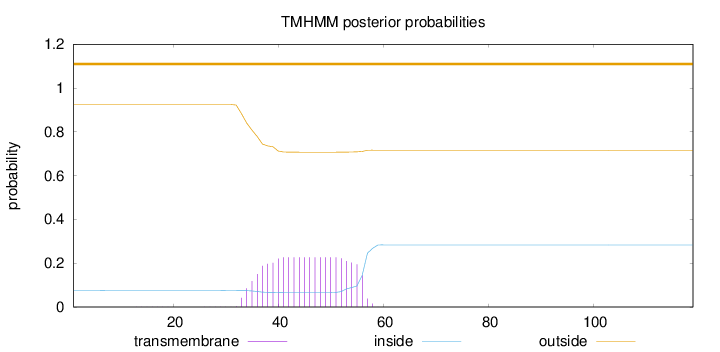
Length:
119
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.73743
Exp number, first 60 AAs:
4.73743
Total prob of N-in:
0.07504
outside
1 - 119
Population Genetic Test Statistics
Pi
223.859597
Theta
202.949129
Tajima's D
0.324332
CLR
4.542336
CSRT
0.461426928653567
Interpretation
Uncertain
