Gene
KWMTBOMO14465
Pre Gene Modal
BGIBMGA000144
Annotation
PREDICTED:_mitogen-activated_protein_kinase_kinase_kinase_kinase_5_isoform_X1_[Bombyx_mori]
Full name
Mitogen-activated protein kinase kinase kinase kinase
Location in the cell
Nuclear Reliability : 3.992
Sequence
CDS
ATGGCGCATAGTGGTGGAGTTTTGAGCTCAGATATATCTCGTAGAAATCCCCAAGATGAGTATGAATTGGTGCAGAGGATCGGTTCTGGAACCTATGGCGATGTATACAAGGCGAAAAGGCTTAACGGCAACGGTGAACTTGCCGCCATCAAGGTGATAAAACTGGAGCCCGGCGACGACTTCGCGATCATCCAACAAGAGATCCTCATGATGAAGGACTGCCGGCATCCGAACATCGTCGCTTATTACGGCTCGTATCTGCGCAGGGATAAACTGTGGATCTCTATGGAGTATTGCGGTGGAGGGAGCTTGCAGGACATCTATCACGTGACTGGGCCATTGACAGAGCTTCAAATAGCCTACATGTGTAGAGAGACCCTTATGGGTTTGACTTATCTACACGGCATGGGGAAGATGCATAGGGACATAAAAGGTGCTAATATCCTGTTGACGGAATGCGGCGATGTTAAGCTTGCAGATTTCGGGGTTTCTGCTCAGATTACGGCCACGATCAACAAGCGGAAGTCCTTCATTGGCACCCCTTATTGGATGGCGCCAGAGGTTGCCGCTGTTGAGAGGAAGGGGGGATACAACCAACTCTGCGATATATGGGCCTGCGGTATAACAGCCATAGAACTAGCGGAACTCCAGCCTCCTATGTTCGAATTGCATCCGATGCGGGTGCTCTTCCTCATGTCGAAGTCCGGCTTCAAGCCGCCGCAGCTGAGGGACAAGGAGCGATGGTCGCAGCTGTTCCACGGGTTCCTGAAGCTGGCGCTCACGAAGAATCCCAAGAAGAGACCGACGGCTCAGAAACTGTTACAGCATCCGTTCTTCCTGCAAGAGATGAGCAAGAGGCTAGCGATAGAGCTGCTGCAGAAGTATTCGAATCCACCGACGCACTGCAACCCGGAGCCGGACGAGGACGCGGGTATATCGAGTGTGCCTCAACGCATAGCGTCGCGCCACACGAACCGAGGGGGGCGCCGGATACTCGCCGAGCGGCTCCGCTCGCCAGGCAACTCCGTCGACAGGCTGCGACCGAGAAGCCTGGTCCCCGAGCCGGCCCCCCCCTCGGACGAGGAGGGCGCCATCACCTTCACCGCGCCCCCCGCCGCCAGGGCTGATACCATCAATAACACTGTGTACCATAGACAATCGAGCGAAGACTGGAGCGTGGCCTCGATCATAAGCTGCCCGAAACACAATCCAATATCTCAGGATCTGTCCTTGGACACGATGCCGAGCAGCGAAAAGAGCCTCTTGCAATATATTGACGAAGAGTTGATGCTCAGGGCAACACTACCGTTGACAGCTGACACGACACTGCCGCAGGCTGGCAACACAGGTCTTTCGTTGCACGACCAACACCTCTCGCAGTGTATACAGTTGAAACAAAACTACTTGAACAATTCAAGTACGAACATTTACCAGAATTTAGCTTCGAATTTTCAAAGTCCGATCGGCACGTCGCGAGATATCGACGAAGACATCTTATACGTTGATCAGTTAGAGGATTCGCCACGGTTGTTGAACGCGAATTGCGAGTGTGGTTTATGTCCGAAGCCAGAGAGGAAAGGCGATAACACTCTGAGCGATTTACAGAAAAGCGTGGCGCCGAAATGCTCGTGCGACTCTTGCAACTCCAACGGCGATATCCTCAGCTATTACAGGAACTGTACTAGCCAGAACTCTGACTCTGGTATCGTTTACTGCGAGAAATGCAACAAGCAGAAGATATCCGTTTCGAACTTCCGAGCTTTACAGATAGCGAACAGCCTCAGCATGGCTGATGACGAAACCCGCAATGGTAGCGTATCGGACGACGACCTCTGTCGGCGGATCGACATGAGCCTCAAGATGGACGAGAAATTCGAGAACAGGAAACGGCACAATCGACACGGCGCAGATACGACGACTTCCGGCATAGACCTGAGTCAGTTCTGTCAGTGCGAGTTTGAGGGTAGGCGTAAAACGTCGTCCGTCGATGAGATATTCAAAATGGTTGAAGAGAAAGACGTCGAGGGCTCGGTTGAGGATGAGGCGAAGGTTAGCCAGCGGCAGAGGAGCTTGAGCGACAGTCAGAGGGATAAAGCGAAAGTCGAGAACAAAGTGTCAGGTCCGGGTACGCCGCCGGTGCCGCCCCGCCGCGCGCGCTCCCGCCGCCTCACCCCTCCGCGCCCCGCACCGAACGGACTCCCCCCCACGCCCAAGGTGCACATGGGGGCTTGCTTCTCCAAGGTGTTCAACGGATGTCCGCTAAGGATAAACTGCACGGCGTCGTGGATACACCCGGAGACGAGAGACCAGCACATACTGATAGGTGCTGAAGAGGGCATCTATACATTGAACCTGAACGAGCTCCACGAGACGGCGATGGACCAGCTCTGCCCCAGGAGGACCATATGGATGCACGTCATCAAAGACGTCCTCATGTCGCTGTCGGGCAAGACGGCGTCTCTGTACCGGCATGAGCTTCTGGCGCTGGCGGGAGGCGCGCGGGGCGGAAGGTCGCTCCGGGTCCTCGCCCCCAAGCTGCTGCCCAGACGGTTCGCCCTCACCACCCGCCTTCCAGACACAAGAGGTTGTACGCGCTGCGCAGTGGCCCGCAATCCCTACAACGGGTACAAGTACCTGTGCGGCGCCACTCCCGCGGGCCTCTTCCTCATGCAGTGGTACGATCCGCTCAGGAAGTTCATGTTGCTCAAAAACATAGAATGTAGTGTACCCTCGCCCTTACCGGTCTTCGAACTGATCATAACCCCGGACTCCGAGTACCCGCTGCTGTGCATCGGCGCCACCAGGAAACCCCTGAGATTGAACCTTATCAACATCAACTCAGGTGCGACGTGGTTCCACTCAGACGAGCTGGAGCCCTGCGTTGGGGGCTCCAACACGGTCCTGCCGCGACCGGAGCGCCTGCACACACTGCGGGCCGTGCACCAACTCAACAAGCACTCGGTGCTAGTGTGCTACGAGAACGTGGTGGACATCATTCCGGTGTTGCCAGCGTCGCTTGACTCGGGCCGGTGGCGCGGGGACGACGACAAGCGGAGGGGGAAGCTACTCTCCAGAATAGAGTTCGACTTTAATATCGACAGTATACTGTGCCTGGCGGACTCTGTCCTGGCGTTCCACCGGCACGGCGTCCAGGGTCGCTCGCTGCGCAACGCTGAGGTCACCCAGGAGATCACGGACCAGTCCAGGGCCTACAGGCTGCTGCCGCACGACAAGGTGGTAGTGCTGGAGTCTCACACTCTGCAGAGCAACACAATGTCGAACGACGAAGGCAACGATCTCTACATACTGGCCGGACACGAGGCTTCATACTAG
Protein
MAHSGGVLSSDISRRNPQDEYELVQRIGSGTYGDVYKAKRLNGNGELAAIKVIKLEPGDDFAIIQQEILMMKDCRHPNIVAYYGSYLRRDKLWISMEYCGGGSLQDIYHVTGPLTELQIAYMCRETLMGLTYLHGMGKMHRDIKGANILLTECGDVKLADFGVSAQITATINKRKSFIGTPYWMAPEVAAVERKGGYNQLCDIWACGITAIELAELQPPMFELHPMRVLFLMSKSGFKPPQLRDKERWSQLFHGFLKLALTKNPKKRPTAQKLLQHPFFLQEMSKRLAIELLQKYSNPPTHCNPEPDEDAGISSVPQRIASRHTNRGGRRILAERLRSPGNSVDRLRPRSLVPEPAPPSDEEGAITFTAPPAARADTINNTVYHRQSSEDWSVASIISCPKHNPISQDLSLDTMPSSEKSLLQYIDEELMLRATLPLTADTTLPQAGNTGLSLHDQHLSQCIQLKQNYLNNSSTNIYQNLASNFQSPIGTSRDIDEDILYVDQLEDSPRLLNANCECGLCPKPERKGDNTLSDLQKSVAPKCSCDSCNSNGDILSYYRNCTSQNSDSGIVYCEKCNKQKISVSNFRALQIANSLSMADDETRNGSVSDDDLCRRIDMSLKMDEKFENRKRHNRHGADTTTSGIDLSQFCQCEFEGRRKTSSVDEIFKMVEEKDVEGSVEDEAKVSQRQRSLSDSQRDKAKVENKVSGPGTPPVPPRRARSRRLTPPRPAPNGLPPTPKVHMGACFSKVFNGCPLRINCTASWIHPETRDQHILIGAEEGIYTLNLNELHETAMDQLCPRRTIWMHVIKDVLMSLSGKTASLYRHELLALAGGARGGRSLRVLAPKLLPRRFALTTRLPDTRGCTRCAVARNPYNGYKYLCGATPAGLFLMQWYDPLRKFMLLKNIECSVPSPLPVFELIITPDSEYPLLCIGATRKPLRLNLININSGATWFHSDELEPCVGGSNTVLPRPERLHTLRAVHQLNKHSVLVCYENVVDIIPVLPASLDSGRWRGDDDKRRGKLLSRIEFDFNIDSILCLADSVLAFHRHGVQGRSLRNAEVTQEITDQSRAYRLLPHDKVVVLESHTLQSNTMSNDEGNDLYILAGHEASY
Summary
Catalytic Activity
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Similarity
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. STE20 subfamily.
Uniprot
EC Number
2.7.11.1
Pubmed
Proteomes
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
PDB
5J5T
E-value=7.83879e-141,
Score=1287
Ontologies
GO
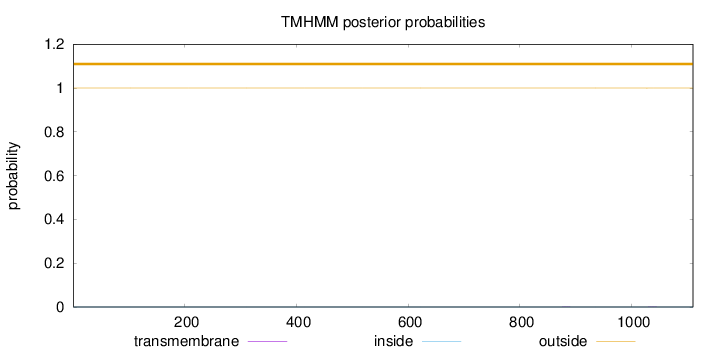
Topology
Length:
1110
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00837
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00009
outside
1 - 1110
Population Genetic Test Statistics
Pi
336.361379
Theta
173.329623
Tajima's D
3.121376
CLR
0.262573
CSRT
0.984650767461627
Interpretation
Uncertain
