Pre Gene Modal
BGIBMGA000138
Annotation
deoxyhypusine_synthase_[Bombyx_mori]
Full name
Probable deoxyhypusine synthase
Location in the cell
PlasmaMembrane Reliability : 1.667
Sequence
CDS
ATGGAAATCCCGTGCAACGTCGAAAAACCTGTTAAAGTAGCCCCAAATATTGCCGTAAATGCGGTTCTGCAACCGAGCCAAGATTTACCTGCAGAAACACCGATAGTCGCGGGCTACGACTGGGAAAATGGAACCGATTACAATAAAATACTCGAATCTTACGCTAACTCCGGATTTCAGGCCACGAATTTCGGTAAAACTGTCGTCGAAATCAATAATATGCTCAAAAGCCGAGCGGTTCCGTTAACCGAAGAAAACAGCGATTGTTATGAAGAAGACCAGTTTATTAAGAAGAAAACTAACTGTACTATATTCCTCGGGTATACTTCTAACATGATATCCTCGGGACTGAGAGACACGATCCGTTTTTTAGTTAAAAATAAGCTTGTGGATGTGATTGTGACTACAGCTGGAGGGATCGAGGAAGACTTGATAAAATGCATCGCTCCGACCTATGTAGGTGATTTTAATTTAAGCGGGAAAGTGTTAAGAACACGAGGTATAAACAGGATAGGCAATTTGTTGGTGCCAAATGATAATTATTGTTTGTTTGAGGAGTGGGTCACTCCATTGTTGGACGAGATGATCTCTGAACAGATGAAAGAGGGTACTGTTTGGAGTCCGTCGCGCATCACAGCCCGGTTAGGGGAGAGGATTAACAATGAGAGCTCAGTGTGCTATTGGGCTTGGAGGAACAACATACCTATATTTAGTCCCGCATTGACTGATGGATCCTTAGGCGACATGATGTACTTCCACTCGTTTAAGCGTCCCGGACTGATCATAGACATATTGGGAGACCTGCGCAGATTGAACACCATGGCCGTCAAAGCCAACAACACTGGAGTCATAATATTAGGTGGGGGTTTGATCAAACATCATATTTGTAACGCAAATCTGATGCGTAATGGTGCTGATTACGCTGTGTACGTGAACACGGCCAGTGAATTTGACGGCAGTGATGCAGGTGCCAGGCCCGACGAAGCAGTCTCGTGGGGCAAAATCAGACCTAATGCGAATCCAGTGAAGCTATATGCAGATGCTACGTTAGTGTTCCCCTTGATCGTAGCTCAAACATTTGCTAAACATCATTTTAGCAGTAAGAAAAATGTTTAA
Protein
MEIPCNVEKPVKVAPNIAVNAVLQPSQDLPAETPIVAGYDWENGTDYNKILESYANSGFQATNFGKTVVEINNMLKSRAVPLTEENSDCYEEDQFIKKKTNCTIFLGYTSNMISSGLRDTIRFLVKNKLVDVIVTTAGGIEEDLIKCIAPTYVGDFNLSGKVLRTRGINRIGNLLVPNDNYCLFEEWVTPLLDEMISEQMKEGTVWSPSRITARLGERINNESSVCYWAWRNNIPIFSPALTDGSLGDMMYFHSFKRPGLIIDILGDLRRLNTMAVKANNTGVIILGGGLIKHHICNANLMRNGADYAVYVNTASEFDGSDAGARPDEAVSWGKIRPNANPVKLYADATLVFPLIVAQTFAKHHFSSKKNV
Summary
Description
Catalyzes the NAD-dependent oxidative cleavage of spermidine and the subsequent transfer of the butylamine moiety of spermidine to the epsilon-amino group of a specific lysine residue of the eIF-5A precursor protein to form the intermediate deoxyhypusine residue.
Catalytic Activity
[eIF5A protein]-L-lysine + spermidine = [eIF5A protein]-deoxyhypusine + propane-1,3-diamine
Cofactor
NAD(+)
Similarity
Belongs to the deoxyhypusine synthase family.
Keywords
Complete proteome
Hypusine biosynthesis
NAD
Reference proteome
Transferase
Feature
chain Probable deoxyhypusine synthase
Uniprot
B5U1G9
A0A0L7LAG2
A0A2A4JHM6
A0A2W1BMV9
A0A2H1W2K7
A0A212ER49
+ More
A0A0N0PA07 A0A0N0PE11 S4P9B0 B0W851 A0A1J1I595 A0A0M9A691 A0A0A1WU90 A0A182IQF1 B4LCB4 W5JCV2 A0A182NIW0 A0A084WML8 A0A154PBP2 R4FP41 B4L933 A0A182X4C3 A0A3B0KQP4 A0A2A3ENU7 A0A1I8MG00 B4J215 A0A182VAX5 Q7Q5B3 A0A182WBW0 A0A0L7R9J5 A0A1B6I1W8 B3M9N0 A0A182MLP7 A0A182UF31 A0A067QIV3 A0A1B6FFZ7 A0A087ZRT5 A0A182I4B8 A0A0M3QWX4 Q16QM1 A0A224XII6 A0A1I8PFH9 B5DP22 Q9VSF4 A0A182JVT6 A0A1A9WLL2 B4H171 B4PGJ5 A0A2J7QSV9 A0A1W4UEB7 A0A1A9ZDX2 B4MKQ6 A0A1A9UKR0 B4QLJ8 A0A182GFB2 B4HJE2 A0A1B6M5G2 A0A0L0CAD5 B3NFW2 A0A1B0C6P8 A0A3Q0JCJ7 A0A1A9XJV6 A0A336M7P4 A0A182Y7X3 D3TQ86 A0A293LNU7 T1J796 E9HAW4 A0A0P5XWB0 A0A0P5LTM4 A0A164ZV65 A0A0N8DMV4 A0A069DT87 A0A2H8TF80 A0A182FFE2 C4WS47 A0A1Y1LH70 A0A182LKL3 A0A0A9XNZ3 A0A2S2QFW8 A0A1E1XRS5 A0A0K8SL72 A0A146KVE2 A0A0K8SL13 A0A023GLK3 A0A146L376 A0A182PRZ2 A0A023F8D9 A0A310SN71 L7M7U6 A0A232EQD1 A0A0P6E565 A0A1B6CH48 A0A0P6DMB5 W8CE17 A0A1E1X4T1 A0A0P5SXR7 A0A1B0GJL8 A0A131XG47 A0A1B6E0I2
A0A0N0PA07 A0A0N0PE11 S4P9B0 B0W851 A0A1J1I595 A0A0M9A691 A0A0A1WU90 A0A182IQF1 B4LCB4 W5JCV2 A0A182NIW0 A0A084WML8 A0A154PBP2 R4FP41 B4L933 A0A182X4C3 A0A3B0KQP4 A0A2A3ENU7 A0A1I8MG00 B4J215 A0A182VAX5 Q7Q5B3 A0A182WBW0 A0A0L7R9J5 A0A1B6I1W8 B3M9N0 A0A182MLP7 A0A182UF31 A0A067QIV3 A0A1B6FFZ7 A0A087ZRT5 A0A182I4B8 A0A0M3QWX4 Q16QM1 A0A224XII6 A0A1I8PFH9 B5DP22 Q9VSF4 A0A182JVT6 A0A1A9WLL2 B4H171 B4PGJ5 A0A2J7QSV9 A0A1W4UEB7 A0A1A9ZDX2 B4MKQ6 A0A1A9UKR0 B4QLJ8 A0A182GFB2 B4HJE2 A0A1B6M5G2 A0A0L0CAD5 B3NFW2 A0A1B0C6P8 A0A3Q0JCJ7 A0A1A9XJV6 A0A336M7P4 A0A182Y7X3 D3TQ86 A0A293LNU7 T1J796 E9HAW4 A0A0P5XWB0 A0A0P5LTM4 A0A164ZV65 A0A0N8DMV4 A0A069DT87 A0A2H8TF80 A0A182FFE2 C4WS47 A0A1Y1LH70 A0A182LKL3 A0A0A9XNZ3 A0A2S2QFW8 A0A1E1XRS5 A0A0K8SL72 A0A146KVE2 A0A0K8SL13 A0A023GLK3 A0A146L376 A0A182PRZ2 A0A023F8D9 A0A310SN71 L7M7U6 A0A232EQD1 A0A0P6E565 A0A1B6CH48 A0A0P6DMB5 W8CE17 A0A1E1X4T1 A0A0P5SXR7 A0A1B0GJL8 A0A131XG47 A0A1B6E0I2
EC Number
2.5.1.46
Pubmed
19121390
26227816
28756777
22118469
26354079
23622113
+ More
25830018 17994087 20920257 23761445 24438588 25315136 12364791 14747013 17210077 24845553 17510324 15632085 23185243 10731132 12537572 17550304 22936249 26483478 26108605 25244985 20353571 21292972 26334808 28004739 20966253 25401762 29209593 26823975 25474469 25576852 28648823 24495485 28503490 28049606
25830018 17994087 20920257 23761445 24438588 25315136 12364791 14747013 17210077 24845553 17510324 15632085 23185243 10731132 12537572 17550304 22936249 26483478 26108605 25244985 20353571 21292972 26334808 28004739 20966253 25401762 29209593 26823975 25474469 25576852 28648823 24495485 28503490 28049606
EMBL
BABH01024231
FJ156236
ACH95798.1
JTDY01001952
KOB72462.1
NWSH01001379
+ More
PCG71471.1 KZ149963 PZC76268.1 ODYU01005924 SOQ47325.1 AGBW02013121 OWR43955.1 KQ458995 KPJ04422.1 KQ460129 KPJ17558.1 GAIX01005581 JAA86979.1 DS231857 EDS38676.1 CVRI01000042 CRK95461.1 KQ435742 KOX76699.1 GBXI01012096 JAD02196.1 CH940647 EDW68759.1 ADMH02001882 ETN60659.1 ATLV01024482 KE525352 KFB51462.1 KQ434869 KZC09253.1 ACPB03017427 GAHY01001064 JAA76446.1 CH933816 EDW17208.1 OUUW01000012 SPP87541.1 KZ288202 PBC33483.1 CH916366 EDV95940.1 AAAB01008960 EAA11387.4 KQ414625 KOC67533.1 GECU01026851 JAS80855.1 CH902618 EDV39036.1 AXCM01000273 KK853297 KDR08786.1 GECZ01020664 JAS49105.1 APCN01002300 CP012525 ALC44933.1 CH477744 EAT36696.1 GFTR01005562 JAW10864.1 CH379067 EDY73273.1 KRT07349.1 AE014296 BT010229 CH479202 EDW30048.1 CM000159 EDW93213.1 NEVH01011219 PNF31653.1 CH963847 EDW72762.2 CM000363 CM002912 EDX09658.1 KMY98314.1 JXUM01000836 JXUM01000837 CH480815 EDW40666.1 GEBQ01008817 JAT31160.1 JRES01000681 KNC29217.1 CH954178 EDV50724.1 JXJN01026846 UFQS01000509 UFQT01000509 SSX04489.1 SSX24853.1 EZ423588 ADD19864.1 GFWV01005673 MAA30403.1 JH431916 GL732613 EFX71102.1 GDIP01066318 JAM37397.1 GDIQ01165834 JAK85891.1 LRGB01000687 KZS16814.1 GDIP01018215 JAM85500.1 GBGD01001933 JAC86956.1 GFXV01000962 MBW12767.1 ABLF02013770 AK340044 BAH70717.1 GEZM01058748 JAV71690.1 GBHO01021147 JAG22457.1 GGMS01006879 MBY76082.1 GFAA01001678 JAU01757.1 GBRD01011826 JAG53998.1 GDHC01019423 JAP99205.1 GBRD01011827 JAG53997.1 GBBM01001450 JAC33968.1 GDHC01015665 JAQ02964.1 GBBI01001147 JAC17565.1 KQ762031 OAD56379.1 GACK01004663 JAA60371.1 NNAY01002808 OXU20532.1 GDIQ01082262 JAN12475.1 GEDC01024540 JAS12758.1 GDIQ01076603 JAN18134.1 GAMC01000976 JAC05580.1 GFAC01004909 JAT94279.1 GDIP01142048 GDIP01140901 JAL62813.1 AJWK01021979 GEFH01003436 JAP65145.1 GEDC01005860 JAS31438.1
PCG71471.1 KZ149963 PZC76268.1 ODYU01005924 SOQ47325.1 AGBW02013121 OWR43955.1 KQ458995 KPJ04422.1 KQ460129 KPJ17558.1 GAIX01005581 JAA86979.1 DS231857 EDS38676.1 CVRI01000042 CRK95461.1 KQ435742 KOX76699.1 GBXI01012096 JAD02196.1 CH940647 EDW68759.1 ADMH02001882 ETN60659.1 ATLV01024482 KE525352 KFB51462.1 KQ434869 KZC09253.1 ACPB03017427 GAHY01001064 JAA76446.1 CH933816 EDW17208.1 OUUW01000012 SPP87541.1 KZ288202 PBC33483.1 CH916366 EDV95940.1 AAAB01008960 EAA11387.4 KQ414625 KOC67533.1 GECU01026851 JAS80855.1 CH902618 EDV39036.1 AXCM01000273 KK853297 KDR08786.1 GECZ01020664 JAS49105.1 APCN01002300 CP012525 ALC44933.1 CH477744 EAT36696.1 GFTR01005562 JAW10864.1 CH379067 EDY73273.1 KRT07349.1 AE014296 BT010229 CH479202 EDW30048.1 CM000159 EDW93213.1 NEVH01011219 PNF31653.1 CH963847 EDW72762.2 CM000363 CM002912 EDX09658.1 KMY98314.1 JXUM01000836 JXUM01000837 CH480815 EDW40666.1 GEBQ01008817 JAT31160.1 JRES01000681 KNC29217.1 CH954178 EDV50724.1 JXJN01026846 UFQS01000509 UFQT01000509 SSX04489.1 SSX24853.1 EZ423588 ADD19864.1 GFWV01005673 MAA30403.1 JH431916 GL732613 EFX71102.1 GDIP01066318 JAM37397.1 GDIQ01165834 JAK85891.1 LRGB01000687 KZS16814.1 GDIP01018215 JAM85500.1 GBGD01001933 JAC86956.1 GFXV01000962 MBW12767.1 ABLF02013770 AK340044 BAH70717.1 GEZM01058748 JAV71690.1 GBHO01021147 JAG22457.1 GGMS01006879 MBY76082.1 GFAA01001678 JAU01757.1 GBRD01011826 JAG53998.1 GDHC01019423 JAP99205.1 GBRD01011827 JAG53997.1 GBBM01001450 JAC33968.1 GDHC01015665 JAQ02964.1 GBBI01001147 JAC17565.1 KQ762031 OAD56379.1 GACK01004663 JAA60371.1 NNAY01002808 OXU20532.1 GDIQ01082262 JAN12475.1 GEDC01024540 JAS12758.1 GDIQ01076603 JAN18134.1 GAMC01000976 JAC05580.1 GFAC01004909 JAT94279.1 GDIP01142048 GDIP01140901 JAL62813.1 AJWK01021979 GEFH01003436 JAP65145.1 GEDC01005860 JAS31438.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000002320 UP000183832 UP000053105 UP000075880 UP000008792 UP000000673 UP000075884 UP000030765 UP000076502 UP000015103 UP000009192 UP000076407 UP000268350 UP000242457 UP000095301 UP000001070 UP000075903 UP000007062 UP000075920 UP000053825 UP000007801 UP000075883 UP000075902 UP000027135 UP000005203 UP000075840 UP000092553 UP000008820 UP000095300 UP000001819 UP000000803 UP000075881 UP000091820 UP000008744 UP000002282 UP000235965 UP000192221 UP000092445 UP000007798 UP000078200 UP000000304 UP000069940 UP000001292 UP000037069 UP000008711 UP000092460 UP000079169 UP000092443 UP000076408 UP000000305 UP000076858 UP000069272 UP000007819 UP000075882 UP000075885 UP000215335 UP000092461
UP000002320 UP000183832 UP000053105 UP000075880 UP000008792 UP000000673 UP000075884 UP000030765 UP000076502 UP000015103 UP000009192 UP000076407 UP000268350 UP000242457 UP000095301 UP000001070 UP000075903 UP000007062 UP000075920 UP000053825 UP000007801 UP000075883 UP000075902 UP000027135 UP000005203 UP000075840 UP000092553 UP000008820 UP000095300 UP000001819 UP000000803 UP000075881 UP000091820 UP000008744 UP000002282 UP000235965 UP000192221 UP000092445 UP000007798 UP000078200 UP000000304 UP000069940 UP000001292 UP000037069 UP000008711 UP000092460 UP000079169 UP000092443 UP000076408 UP000000305 UP000076858 UP000069272 UP000007819 UP000075882 UP000075885 UP000215335 UP000092461
Interpro
IPR036982
Deoxyhypusine_synthase_sf
+ More
IPR029035 DHS-like_NAD/FAD-binding_dom
IPR002773 Deoxyhypusine_synthase
IPR006722 Sedlin
IPR011012 Longin-like_dom_sf
IPR002347 SDR_fam
IPR036291 NAD(P)-bd_dom_sf
IPR019013 Vma21
IPR026904 GidA-assoc_3
IPR004416 MnmG
IPR020595 MnmG-rel_CS
IPR002218 MnmG-rel
IPR036188 FAD/NAD-bd_sf
IPR029035 DHS-like_NAD/FAD-binding_dom
IPR002773 Deoxyhypusine_synthase
IPR006722 Sedlin
IPR011012 Longin-like_dom_sf
IPR002347 SDR_fam
IPR036291 NAD(P)-bd_dom_sf
IPR019013 Vma21
IPR026904 GidA-assoc_3
IPR004416 MnmG
IPR020595 MnmG-rel_CS
IPR002218 MnmG-rel
IPR036188 FAD/NAD-bd_sf
Gene 3D
ProteinModelPortal
B5U1G9
A0A0L7LAG2
A0A2A4JHM6
A0A2W1BMV9
A0A2H1W2K7
A0A212ER49
+ More
A0A0N0PA07 A0A0N0PE11 S4P9B0 B0W851 A0A1J1I595 A0A0M9A691 A0A0A1WU90 A0A182IQF1 B4LCB4 W5JCV2 A0A182NIW0 A0A084WML8 A0A154PBP2 R4FP41 B4L933 A0A182X4C3 A0A3B0KQP4 A0A2A3ENU7 A0A1I8MG00 B4J215 A0A182VAX5 Q7Q5B3 A0A182WBW0 A0A0L7R9J5 A0A1B6I1W8 B3M9N0 A0A182MLP7 A0A182UF31 A0A067QIV3 A0A1B6FFZ7 A0A087ZRT5 A0A182I4B8 A0A0M3QWX4 Q16QM1 A0A224XII6 A0A1I8PFH9 B5DP22 Q9VSF4 A0A182JVT6 A0A1A9WLL2 B4H171 B4PGJ5 A0A2J7QSV9 A0A1W4UEB7 A0A1A9ZDX2 B4MKQ6 A0A1A9UKR0 B4QLJ8 A0A182GFB2 B4HJE2 A0A1B6M5G2 A0A0L0CAD5 B3NFW2 A0A1B0C6P8 A0A3Q0JCJ7 A0A1A9XJV6 A0A336M7P4 A0A182Y7X3 D3TQ86 A0A293LNU7 T1J796 E9HAW4 A0A0P5XWB0 A0A0P5LTM4 A0A164ZV65 A0A0N8DMV4 A0A069DT87 A0A2H8TF80 A0A182FFE2 C4WS47 A0A1Y1LH70 A0A182LKL3 A0A0A9XNZ3 A0A2S2QFW8 A0A1E1XRS5 A0A0K8SL72 A0A146KVE2 A0A0K8SL13 A0A023GLK3 A0A146L376 A0A182PRZ2 A0A023F8D9 A0A310SN71 L7M7U6 A0A232EQD1 A0A0P6E565 A0A1B6CH48 A0A0P6DMB5 W8CE17 A0A1E1X4T1 A0A0P5SXR7 A0A1B0GJL8 A0A131XG47 A0A1B6E0I2
A0A0N0PA07 A0A0N0PE11 S4P9B0 B0W851 A0A1J1I595 A0A0M9A691 A0A0A1WU90 A0A182IQF1 B4LCB4 W5JCV2 A0A182NIW0 A0A084WML8 A0A154PBP2 R4FP41 B4L933 A0A182X4C3 A0A3B0KQP4 A0A2A3ENU7 A0A1I8MG00 B4J215 A0A182VAX5 Q7Q5B3 A0A182WBW0 A0A0L7R9J5 A0A1B6I1W8 B3M9N0 A0A182MLP7 A0A182UF31 A0A067QIV3 A0A1B6FFZ7 A0A087ZRT5 A0A182I4B8 A0A0M3QWX4 Q16QM1 A0A224XII6 A0A1I8PFH9 B5DP22 Q9VSF4 A0A182JVT6 A0A1A9WLL2 B4H171 B4PGJ5 A0A2J7QSV9 A0A1W4UEB7 A0A1A9ZDX2 B4MKQ6 A0A1A9UKR0 B4QLJ8 A0A182GFB2 B4HJE2 A0A1B6M5G2 A0A0L0CAD5 B3NFW2 A0A1B0C6P8 A0A3Q0JCJ7 A0A1A9XJV6 A0A336M7P4 A0A182Y7X3 D3TQ86 A0A293LNU7 T1J796 E9HAW4 A0A0P5XWB0 A0A0P5LTM4 A0A164ZV65 A0A0N8DMV4 A0A069DT87 A0A2H8TF80 A0A182FFE2 C4WS47 A0A1Y1LH70 A0A182LKL3 A0A0A9XNZ3 A0A2S2QFW8 A0A1E1XRS5 A0A0K8SL72 A0A146KVE2 A0A0K8SL13 A0A023GLK3 A0A146L376 A0A182PRZ2 A0A023F8D9 A0A310SN71 L7M7U6 A0A232EQD1 A0A0P6E565 A0A1B6CH48 A0A0P6DMB5 W8CE17 A0A1E1X4T1 A0A0P5SXR7 A0A1B0GJL8 A0A131XG47 A0A1B6E0I2
PDB
1RQD
E-value=4.17666e-125,
Score=1146
Ontologies
GO
PANTHER
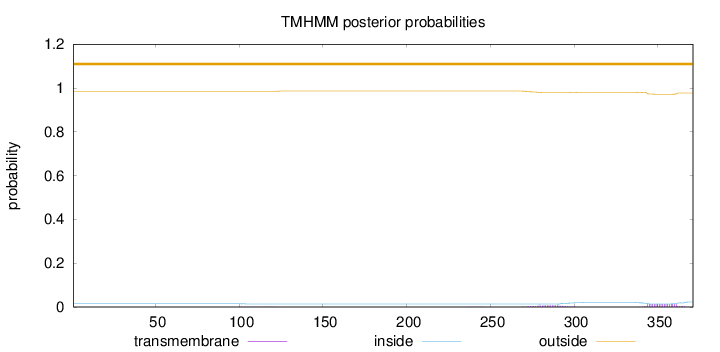
Topology
Length:
371
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.43956
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01476
outside
1 - 371
Population Genetic Test Statistics
Pi
240.916608
Theta
191.929157
Tajima's D
0.637732
CLR
42.847604
CSRT
0.556172191390431
Interpretation
Uncertain
