Gene
KWMTBOMO14454 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000136
Annotation
PREDICTED:_MAP/microtubule_affinity-regulating_kinase_3-like_isoform_X5_[Bombyx_mori]
Full name
Non-specific serine/threonine protein kinase
Location in the cell
Nuclear Reliability : 4.379
Sequence
CDS
ATGAAAATGCTTGATCATCCGAACATTGTGAAGCTCTTCCAAGTTATAGAGACTGAAAAGACACTTTACCTGGTCATGGAATATGCTTCGGGTGGAGAGGTGTTCGACTATTTAGTGCTACACGGCCGCATGAAGGAGAAGGAGGCCCGTGCTAAGTTCAGGCAGATAGTTTCGGCCGTCCAATACTGCCACCAGAAACGGATCATACATAGGGATCTCAAGGCCGAGAATCTTTTATTAGACGGCGAGATGAACATTAAGATTGCCGATTTCGGTTTCTCTAACGAGTTCACGCCGGGAGCGAAGCTGGACACCTTCTGTGGCAGTCCGCCCTATGCAGCCCCTGAACTGTTCCAAGGCAAGAAATACGACGGTCCCGAGGTCGACGTGTGGTCTCTGGGCGTGATCCTCTACACGCTGGTCTCCGGCTCGCTGCCCTTCGACGGCTCCACGCTCCGGGAGCTACGCGAGAGGGTGCTGCGAGGGAAATACAGAATACCGTTCTACATGTCAACGGACTGCGAGAATCTTTTAAAGAAATTCCTCGTCCTGAATCCTGCGAAGCGTGCGTCCCTAGAGAGCATAATGCGAGACAAATGGATGAACACCGGATACGAGGAGGACGAACTCAGGCCCTACGTCGAGCCGCAGCAGGACTTCAAGGACCACAAGCGGATAGAGGCGTTAGTCTGCCTGGGCTACAACAGACAAGAGATTGAATACTCCCTGGCTGAAGCGAAATACGATGACGTATTCGCCACATACTTACTGCTCGGAAGGAAAAGTACCGATCCGGAGTCCGATGGCAGTCGCTCGGGCTCGTCTCTGTCCCTACGGACGTCGGCCGCCGCCAACCAGACCCAGCAGCAGCAGCAGACCAACTCGCACGCAGGGTCGGGCTCGCCGTCGCACCGCGGCGTGCAGCGCTCCATCTCGGCGAGTGCGAGGCCTCAGCCCTCGCGCCGAGCCTCCTCCGGCGCTGAGGTCCTCAGGACCCAAGTTGGTACGACGACGACCGCGTCAAACAGTACGACTTCAACGGCGACCACTACCGCTTCTAATTTCAAAAGACAGAACACAATCGACTCGGCGTCGATCAAAGAAAACACAGCACGGATCGCTCAAATGCAAGTACAAAGCACGACTCGTCCGACTAGCGCCCAGCCCAAGCTAGAGCCGCGGTCTCGCACGTTGACGGGTGGTGCGCGGCGCTCCCACGCCAACTACGAGCCGCCCGCGCACGCGCCCAAGCTGTCGCCGCCGCACGATGCGCCCAGTTTGAACACGCAGTTGACAGTGACTGGCGCCAGTACTGGCACCGGCTCGGCTGCGAGTGCGGCCAGACGCGAGTTCCCCAACCAGCTCGTCTTCCCCAGGAACGTACCTTCGCGCTCCACCTTCCACTCAGGGCAGACGCGGTCGCGCGGGGCGGGCGGCGCGGCGGGGTACGGGGAGGGTGGCTCCCCGCACCAACCACGGCCCTCATTCTTCTCCAAACTGTCCTCGCGGTTCTCTAAACGTTTGTCGGTGCTGGGAGAGTCGACTGGTCGTCAGAAACCTCGTATTTTTTCCTCTCTGGACATACAGCGGAGACAAGGAAACGAGGAACAAGTGAAGCCCAGGGTGCTTCGCTTCACGTGGAGCATGAAGACGACCTCGTCGCGGGACCCGAATGAGATCATGGCTGAGATCAGGAAGGTGCTCGATGCGAACAACTGCGATTACGAGCAGCGCGAGAGGTTCTTATTGCTGTGCGTGCACGGCGACCCCAACGCGGACTCGCTGGTGCAGTGGGAGATCGAGGTCTGCAAGCTGCCGCGCCTCTCGCTCAACGGGGTGCGCTTCAAGAGGATATCCGGAACAAGCATCGGCTTCAAGAACATTGCGTCAAAGATAGCGAACGAACTCAAGCTCCTGCGCCTCTCCGCCGAAGACATGGTGCTGATCGCCAACGGGGGCCAGCGGACGTCCTCCGGCGCGCTGGCTCACTTCCTGGCCGCGTACAGTCCGGCCGAGCTGTCTCTGGCCCGCAGCGACTACCTGCGGCCGCCGGGCTCCCCGCCCCGCCTCCACCCCCGCTCGGCCCTGTCCCTGCGCGCCGACAGCGCCGACGTGCCCTCCCCCTACCGCGTGGCTTACTCCAGCACGCACACGCTGCGCCGACAGCCCCGCGCCCTCTCGCCCCCCACACCCCCGTCACCGTGCGACTGTCTGCCCCTCGAGCCGATCCCCCCTCCTGAGCTGTTCGATTTTTAA
Protein
MKMLDHPNIVKLFQVIETEKTLYLVMEYASGGEVFDYLVLHGRMKEKEARAKFRQIVSAVQYCHQKRIIHRDLKAENLLLDGEMNIKIADFGFSNEFTPGAKLDTFCGSPPYAAPELFQGKKYDGPEVDVWSLGVILYTLVSGSLPFDGSTLRELRERVLRGKYRIPFYMSTDCENLLKKFLVLNPAKRASLESIMRDKWMNTGYEEDELRPYVEPQQDFKDHKRIEALVCLGYNRQEIEYSLAEAKYDDVFATYLLLGRKSTDPESDGSRSGSSLSLRTSAAANQTQQQQQTNSHAGSGSPSHRGVQRSISASARPQPSRRASSGAEVLRTQVGTTTTASNSTTSTATTTASNFKRQNTIDSASIKENTARIAQMQVQSTTRPTSAQPKLEPRSRTLTGGARRSHANYEPPAHAPKLSPPHDAPSLNTQLTVTGASTGTGSAASAARREFPNQLVFPRNVPSRSTFHSGQTRSRGAGGAAGYGEGGSPHQPRPSFFSKLSSRFSKRLSVLGESTGRQKPRIFSSLDIQRRQGNEEQVKPRVLRFTWSMKTTSSRDPNEIMAEIRKVLDANNCDYEQRERFLLLCVHGDPNADSLVQWEIEVCKLPRLSLNGVRFKRISGTSIGFKNIASKIANELKLLRLSAEDMVLIANGGQRTSSGALAHFLAAYSPAELSLARSDYLRPPGSPPRLHPRSALSLRADSADVPSPYRVAYSSTHTLRRQPRALSPPTPPSPCDCLPLEPIPPPELFDF
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Uniprot
A0A2A4K5G1
A0A2A4K436
A0A2A4K421
A0A2A4K5G8
A0A2A4K5R1
A0A2A4K4L1
+ More
A0A2A4K4Q8 A0A1Y1MCF9 A0A1Y1MFA2 A0A1Y1MAG8 A0A1Y1MDQ7 A0A139WKE6 A0A224X7R3 A0A069DWX6 A0A139WKN2 A0A2A4K523 A0A139WKM7 A0A1I8PKX9 A0A1I8PKR7 A0A1I8N449 T1PE90 B3MHB1 A0A0Q9XI31 A0A0R3NSN4 A0A0Q9W3X6 A0A1I8PKZ0 A0A0Q9W4L7 A0A0Q9W403 A0A0R3NTE4 A0A1I8PKY4 A0A1W4V4K6 A0A0Q9WRA8 A0A195EYD1 A0A139WKC9 A0A0Q9WC24 B4LNW1 A0A0Q9XIQ8 A0A1I8PKV9 A0A1I8PKX0 A0A0Q9WGE2 A0A0Q9W4Z7 A0A0Q9XM92 A0A0Q9XMA6 A0A1W4V441 A0A0Q9X928 A0A0Q9XLQ6 A0A0T6BGE6 A0A336LST4 A0A1W4V4J6 A0A0Q9X9W0 A0A0Q9X913 V5FZW2 V5GGV9 V5GSL6 A0A0Q9WDC6 A1ZBL7 A0A0Q9XGB3 A0A1B6CAK5
A0A2A4K4Q8 A0A1Y1MCF9 A0A1Y1MFA2 A0A1Y1MAG8 A0A1Y1MDQ7 A0A139WKE6 A0A224X7R3 A0A069DWX6 A0A139WKN2 A0A2A4K523 A0A139WKM7 A0A1I8PKX9 A0A1I8PKR7 A0A1I8N449 T1PE90 B3MHB1 A0A0Q9XI31 A0A0R3NSN4 A0A0Q9W3X6 A0A1I8PKZ0 A0A0Q9W4L7 A0A0Q9W403 A0A0R3NTE4 A0A1I8PKY4 A0A1W4V4K6 A0A0Q9WRA8 A0A195EYD1 A0A139WKC9 A0A0Q9WC24 B4LNW1 A0A0Q9XIQ8 A0A1I8PKV9 A0A1I8PKX0 A0A0Q9WGE2 A0A0Q9W4Z7 A0A0Q9XM92 A0A0Q9XMA6 A0A1W4V441 A0A0Q9X928 A0A0Q9XLQ6 A0A0T6BGE6 A0A336LST4 A0A1W4V4J6 A0A0Q9X9W0 A0A0Q9X913 V5FZW2 V5GGV9 V5GSL6 A0A0Q9WDC6 A1ZBL7 A0A0Q9XGB3 A0A1B6CAK5
EC Number
2.7.11.1
Pubmed
EMBL
NWSH01000151
PCG79013.1
PCG79017.1
PCG79011.1
PCG79023.1
PCG79012.1
+ More
PCG79009.1 PCG79016.1 GEZM01037681 JAV81985.1 GEZM01037679 JAV81987.1 GEZM01037678 JAV81988.1 GEZM01037680 JAV81986.1 KQ971328 KYB28374.1 GFTR01008040 JAW08386.1 GBGD01000519 JAC88370.1 KYB28375.1 PCG79018.1 KYB28377.1 KA646999 AFP61628.1 CH902619 EDV36888.2 CH933808 KRG04932.1 CM000071 KRT02061.1 CH940648 KRF79801.1 KRF79794.1 KRF79802.1 KRT02051.1 CH963850 KRF98141.1 KQ981906 KYN33228.1 KYB28376.1 KRF79799.1 EDW61130.2 KRG04946.1 KRF79797.1 KRF79798.1 KRG04931.1 KRG04947.1 KRG04934.1 KRG04936.1 LJIG01000519 KRT86421.1 UFQT01000094 SSX19649.1 KRG04933.1 KRG04939.1 GALX01005231 JAB63235.1 GALX01005237 JAB63229.1 GALX01005233 JAB63233.1 KRF79793.1 AE013599 AAS64800.1 KRG04938.1 GEDC01026805 GEDC01024913 JAS10493.1 JAS12385.1
PCG79009.1 PCG79016.1 GEZM01037681 JAV81985.1 GEZM01037679 JAV81987.1 GEZM01037678 JAV81988.1 GEZM01037680 JAV81986.1 KQ971328 KYB28374.1 GFTR01008040 JAW08386.1 GBGD01000519 JAC88370.1 KYB28375.1 PCG79018.1 KYB28377.1 KA646999 AFP61628.1 CH902619 EDV36888.2 CH933808 KRG04932.1 CM000071 KRT02061.1 CH940648 KRF79801.1 KRF79794.1 KRF79802.1 KRT02051.1 CH963850 KRF98141.1 KQ981906 KYN33228.1 KYB28376.1 KRF79799.1 EDW61130.2 KRG04946.1 KRF79797.1 KRF79798.1 KRG04931.1 KRG04947.1 KRG04934.1 KRG04936.1 LJIG01000519 KRT86421.1 UFQT01000094 SSX19649.1 KRG04933.1 KRG04939.1 GALX01005231 JAB63235.1 GALX01005237 JAB63229.1 GALX01005233 JAB63233.1 KRF79793.1 AE013599 AAS64800.1 KRG04938.1 GEDC01026805 GEDC01024913 JAS10493.1 JAS12385.1
Proteomes
PRIDE
Interpro
ProteinModelPortal
A0A2A4K5G1
A0A2A4K436
A0A2A4K421
A0A2A4K5G8
A0A2A4K5R1
A0A2A4K4L1
+ More
A0A2A4K4Q8 A0A1Y1MCF9 A0A1Y1MFA2 A0A1Y1MAG8 A0A1Y1MDQ7 A0A139WKE6 A0A224X7R3 A0A069DWX6 A0A139WKN2 A0A2A4K523 A0A139WKM7 A0A1I8PKX9 A0A1I8PKR7 A0A1I8N449 T1PE90 B3MHB1 A0A0Q9XI31 A0A0R3NSN4 A0A0Q9W3X6 A0A1I8PKZ0 A0A0Q9W4L7 A0A0Q9W403 A0A0R3NTE4 A0A1I8PKY4 A0A1W4V4K6 A0A0Q9WRA8 A0A195EYD1 A0A139WKC9 A0A0Q9WC24 B4LNW1 A0A0Q9XIQ8 A0A1I8PKV9 A0A1I8PKX0 A0A0Q9WGE2 A0A0Q9W4Z7 A0A0Q9XM92 A0A0Q9XMA6 A0A1W4V441 A0A0Q9X928 A0A0Q9XLQ6 A0A0T6BGE6 A0A336LST4 A0A1W4V4J6 A0A0Q9X9W0 A0A0Q9X913 V5FZW2 V5GGV9 V5GSL6 A0A0Q9WDC6 A1ZBL7 A0A0Q9XGB3 A0A1B6CAK5
A0A2A4K4Q8 A0A1Y1MCF9 A0A1Y1MFA2 A0A1Y1MAG8 A0A1Y1MDQ7 A0A139WKE6 A0A224X7R3 A0A069DWX6 A0A139WKN2 A0A2A4K523 A0A139WKM7 A0A1I8PKX9 A0A1I8PKR7 A0A1I8N449 T1PE90 B3MHB1 A0A0Q9XI31 A0A0R3NSN4 A0A0Q9W3X6 A0A1I8PKZ0 A0A0Q9W4L7 A0A0Q9W403 A0A0R3NTE4 A0A1I8PKY4 A0A1W4V4K6 A0A0Q9WRA8 A0A195EYD1 A0A139WKC9 A0A0Q9WC24 B4LNW1 A0A0Q9XIQ8 A0A1I8PKV9 A0A1I8PKX0 A0A0Q9WGE2 A0A0Q9W4Z7 A0A0Q9XM92 A0A0Q9XMA6 A0A1W4V441 A0A0Q9X928 A0A0Q9XLQ6 A0A0T6BGE6 A0A336LST4 A0A1W4V4J6 A0A0Q9X9W0 A0A0Q9X913 V5FZW2 V5GGV9 V5GSL6 A0A0Q9WDC6 A1ZBL7 A0A0Q9XGB3 A0A1B6CAK5
PDB
2QNJ
E-value=1.20625e-131,
Score=1206
Ontologies
GO
GO:0004674
GO:0000226
GO:0030010
GO:0005524
GO:0051124
GO:0050321
GO:0045172
GO:0030860
GO:0016323
GO:0061174
GO:0005938
GO:0007298
GO:0045169
GO:0016325
GO:0007317
GO:0007294
GO:0035331
GO:0090176
GO:0001737
GO:0001932
GO:0031594
GO:0030709
GO:0007318
GO:0019730
GO:0005737
GO:0035556
GO:0009798
GO:0030707
GO:0006468
GO:0009948
GO:0009994
GO:0045451
GO:0004672
GO:0043631
GO:0004652
GO:0005634
GO:0005507
GO:0004713
GO:0016491
GO:0015930
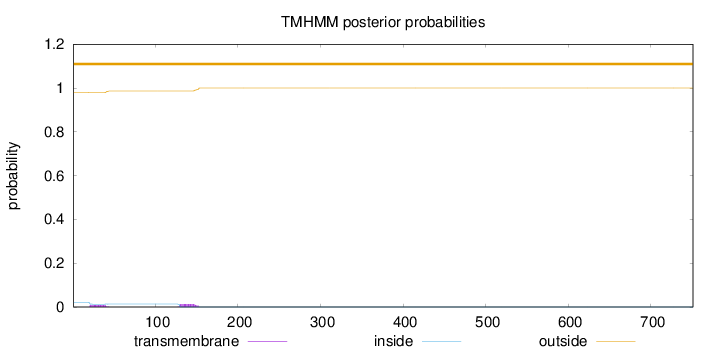
Topology
Length:
751
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.46162
Exp number, first 60 AAs:
0.17413
Total prob of N-in:
0.02111
outside
1 - 751
Population Genetic Test Statistics
Pi
239.791996
Theta
194.744896
Tajima's D
0.827876
CLR
0.304156
CSRT
0.614169291535423
Interpretation
Uncertain
