Gene
KWMTBOMO14447 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000132
Annotation
PREDICTED:_neutral_alpha-glucosidase_AB-like_isoform_X2_[Bombyx_mori]
Location in the cell
Peroxisomal Reliability : 0.836
Sequence
CDS
ATGAAGGTCTGGGCCTTAGTCCTGGTCGCGGCATTTGTGATTATCGGCATTTCTGCGGTAGACAAAAATAAATTTAAAACATGCAGTCAGTCCGGCTTTTGTAAAAGGCTCCGCCCTTTCAAACCAGAAAAATCTCAGTATAGTTTGAATTTGGACAGTATTTTAGTTCACGGCAACGTTCTATCCGCTGAGGTGGTTACCATTGACGCTGCGGATGAAAAGCGAACAGTTTTGTGGAAATATGCCTTCAAGTTATCAGCGCTGGCCGATGGCACGTTCCGTGTGGAGCTCGATGAAGCGGATCCTCTATACCCAAGATACAGGACGCAGCTGGCGTTGGATGGGGAACCGAAGGCAGACGGATTAAAACTGATTTCGAAAGACGATGGAAAAGTTATTCTCGTCAACACGCAAGGGCATAAAGTCATCATAACATCCGAACCTTTGAAACTCGAATTCTTGGACCAGAATGGAGAAGTAGCGGTCGTTCTCAACGAGAACTCTCAGCTGCTCGTGGAGCCTCTAAGGGCGAGAAGGGAAAAGGAGGAAGGAACTTGGAGTGAGACCTTCCAGTCCCATCACGACTCCAAACCTCGGGGCAACGAAGCAGTTTCGCTCGACGTTGTCTTCCCTGATGCGGATCACGTCTACGGTATACCGGAGCACACGGACGGTTTGGCCCTGAAGACGACCACGTCCGGCGACCCGTACAGGCTGTACAACCTGGACGTCTTCGAATACATATTGGACAGCAGGATGGCGATATACGGCTCGGTCCCGGTTATGTACGCACACGGCCCGAAACGGACCGTGGGCGTGTTCTGGCACAACTCCGCCGAGACCTGGATCGATGTGGTCAACTACGGGGAAGGGAACGTGGTCTCGTCGCTCGTCAACCTGGTCACCGGGGGACAGAAGAAACGCGTCGATGCAAGGTTCCTGAGCGAGTCCGGCATCGTGGACATGTTCGTGCTGATGGGCTCCACCCCCGGGGACGCGTTCCGGCAGTACACCGCGCTCACCGGGACCACGCCGCTGCCGCCAAAATTCTCGCTGGGCTACCACCAGTGCCGCTGGAACTACATGGACGAGGCGGACGTGCGCTCCGTCGACGAGAACTTTGACGTGCACGACATCCCCATGGACTCCATCTGGCTGGACATCGAGTACACCAACAAGAAGATGTACTTCACGTGGGATGTGGTCAAGTTTCCGCATCCGGCGGAAATGGTGGCCAACCTGACGGCCAAAGGAAGGAAGATGGTTGTCATCGTGGATCCGCACATCAAACGCGAACCTGGATATTTCTTACACGAGGACGCGACTGAATTAGGGTACTACGTCAAGGACAGAGACGGCAAGGATTATGAGGGTTGGTGCTGGCCTGGCTCGTCGTCGTACCCGGACTTCTTCAACCCGGTGGTCAGCCGGTACTACTCGGACCGGTACAGGTTCGAGAACTTCCCCGGGACTAGCAAGGACGTCCACCTGTGGAACGATATGAACGAGCCCAGTGTCTTCAACGGTCCAGAGGTCACCATGCCCAAGGACTGTCGCCACTACAAGCCGCCTCAGGATGGTCTAGAGGGGCTGGCGGCTTACTGGGAGCATAGGCACGTGCACAACGAATACGGGCTGTGGAACCTGCGCGCCACCAACACGGGGCTGCTGGACCGCGCCGACGGCGTCTACAGACCCTTCCTGCTCACGAGGGCCGTGTTCGCAGGCACCCAGAGATACTCTGCGGTATGGACAGGTGACAATACGGCGGAGTGGTCGTTCCTTGCGGCTTCCGTGCCGATGTGCTTGTCACTGGCAATAGCCGGGAACAGTTTCTGCGGTTCTGACGTCGGAGGATTCTTCAAGTATCCCGAAGCTGAGCTCATGACGAGGTGGTACCAGGCGGCCGCGTTCCAGCCGTTCTTCCGGGCCCACTCGCACATCGAGACCAAGCGGCGCGAGCCCTGGCTGTACCCGGCCGCCACCACCGCGCTCATCCGGGACGCCAACCGCAAGAGATACGCGCTGCTCGACTTCTGGTACACTCTGTTCTACGAGCACACGGTGGACGGGCTGCCGGTCATGAGGCCCCTGTTCCAGCACTACCCCGACGAGCCGGCCACCTACCCCATCGACGACCAGTACCTGCTCGGGGACGCGCTGCTGGTGAAGCCGGTGACGACGGCTGGGGCGAGCTCCGTGGAGGTATACCTGCCCGGGGACGGCCCGTGGTACGACGTGGACACTTACGTGCCGCACAAAGGACCCCGCATCACGCAGCCGGTCACCATCTCCAAGATCCCGGTGTACCAGCGCGGCGGAACCATAGTGCCGCGCCGGGAGCGCGTGCGACGCTCCTCCGCGCTGATGGCTGACGACCCCTTCACCCTGGTGGTCGCGCTCGACGCTAACAACTCTGCTCGCGGCTCTCTGTACATCGACGACGGCGAAACCTACGAGTACAAGAACAATAAGTACATCTACGCCAAGATCGAGTTCGGCCCCGACGAGATGACGTACACGCTCGCGAGCGAGGGCGCCCAGTACCCGACCGGGGCGTGGCTGGAGCGCGTGGTGGTGGCCGGCATCAGGGCGGCGCCGCGGAGTGCCCGGCTGCAGCAGGACGGGCGGCAGACGGCGCTGCAGATGACGCTGCACAAGGGCAACGACGTGCTGGTGCTGAGGAAGCCGGCCGCTAGCATGGCCAGACCCTGGACCATCACCTTCGCCTACTGA
Protein
MKVWALVLVAAFVIIGISAVDKNKFKTCSQSGFCKRLRPFKPEKSQYSLNLDSILVHGNVLSAEVVTIDAADEKRTVLWKYAFKLSALADGTFRVELDEADPLYPRYRTQLALDGEPKADGLKLISKDDGKVILVNTQGHKVIITSEPLKLEFLDQNGEVAVVLNENSQLLVEPLRARREKEEGTWSETFQSHHDSKPRGNEAVSLDVVFPDADHVYGIPEHTDGLALKTTTSGDPYRLYNLDVFEYILDSRMAIYGSVPVMYAHGPKRTVGVFWHNSAETWIDVVNYGEGNVVSSLVNLVTGGQKKRVDARFLSESGIVDMFVLMGSTPGDAFRQYTALTGTTPLPPKFSLGYHQCRWNYMDEADVRSVDENFDVHDIPMDSIWLDIEYTNKKMYFTWDVVKFPHPAEMVANLTAKGRKMVVIVDPHIKREPGYFLHEDATELGYYVKDRDGKDYEGWCWPGSSSYPDFFNPVVSRYYSDRYRFENFPGTSKDVHLWNDMNEPSVFNGPEVTMPKDCRHYKPPQDGLEGLAAYWEHRHVHNEYGLWNLRATNTGLLDRADGVYRPFLLTRAVFAGTQRYSAVWTGDNTAEWSFLAASVPMCLSLAIAGNSFCGSDVGGFFKYPEAELMTRWYQAAAFQPFFRAHSHIETKRREPWLYPAATTALIRDANRKRYALLDFWYTLFYEHTVDGLPVMRPLFQHYPDEPATYPIDDQYLLGDALLVKPVTTAGASSVEVYLPGDGPWYDVDTYVPHKGPRITQPVTISKIPVYQRGGTIVPRRERVRRSSALMADDPFTLVVALDANNSARGSLYIDDGETYEYKNNKYIYAKIEFGPDEMTYTLASEGAQYPTGAWLERVVVAGIRAAPRSARLQQDGRQTALQMTLHKGNDVLVLRKPAASMARPWTITFAY
Summary
Similarity
Belongs to the glycosyl hydrolase 31 family.
Uniprot
L8B8U1
H9ISA5
L8B8V1
A0A0N0PA04
A0A212F746
A0A345W861
+ More
A0A2H1VQT4 A0A139WL23 A0A2A3EPD6 A0A088A7S9 A0A2J7RPJ2 A0A154PM08 A0A067QY16 A0A0N0U565 A0A026WK76 A0A3L8DZF9 U5EKN8 A0A0L7RBC3 A0A336LLR5 A0A1L8DQZ3 A0A1L8DQT5 A0A1L8DQW6 E2BII6 A0A1L8DQR3 A0A1L8DR57 A0A1L8DQT4 A0A195F510 A0A1Y1LFQ8 A0A1J1HNH3 F4W549 A0A195D9U3 A0A158NDK9 A0A195BQQ2 A0A151XEG8 A0A182JCA9 E9IU82 Q16SF9 V5GYU2 A0A1B0ETK8 A0A1S6J0X3 A0A1B6GNL0 A0A2K8FTL6 E2A0I0 A0A1I8P7N9 B4NDN1 T1PHL5 A0A1I8MLM4 A0A232F1E1 A0A084WFF5 A0A224X6Q8 A0A0L0BRC7 A0A2K8JWU1 A0A0C9QW93 A0A3B0JWU7 A0A1A9X589 B0WQR9 A0A182N0N9 A0A1Q3FD85 Q29JE9 T1IAP9 A0A0A9X7R6 B4PZ50 B3NYF5 A0A0A9X6S5 A0A0K8TJ80 B4M886 A0A182QSP4 A0A0K8T7J7 A0A126CR36 A0A1W4V862 A0A182URY2 A0A226ERG3 A0A1B6JRL3 A0A182M5G9 B4L1Z6 A0A0K8V6T5 A0A182WZ94 A0A182XYB2 Q7KMM4 B4JNR8 A0A182R253 A0A182PUA8 Q7Q4V0 A0A1A9UUR7 A0A182HMR5 W8C608 A0A182VVG0 A0A0M4F9S7 A0A3P9B7H3 A0A182TVW1 A0A3B5A1R0 A0A2M3YZB8 A0A2H8TSQ9 A0A1B0G4M6 A0A3Q2HNI5 J9K948 A0A2S2NLM4 A0A1B0AX59 A0A3Q2ZSP1
A0A2H1VQT4 A0A139WL23 A0A2A3EPD6 A0A088A7S9 A0A2J7RPJ2 A0A154PM08 A0A067QY16 A0A0N0U565 A0A026WK76 A0A3L8DZF9 U5EKN8 A0A0L7RBC3 A0A336LLR5 A0A1L8DQZ3 A0A1L8DQT5 A0A1L8DQW6 E2BII6 A0A1L8DQR3 A0A1L8DR57 A0A1L8DQT4 A0A195F510 A0A1Y1LFQ8 A0A1J1HNH3 F4W549 A0A195D9U3 A0A158NDK9 A0A195BQQ2 A0A151XEG8 A0A182JCA9 E9IU82 Q16SF9 V5GYU2 A0A1B0ETK8 A0A1S6J0X3 A0A1B6GNL0 A0A2K8FTL6 E2A0I0 A0A1I8P7N9 B4NDN1 T1PHL5 A0A1I8MLM4 A0A232F1E1 A0A084WFF5 A0A224X6Q8 A0A0L0BRC7 A0A2K8JWU1 A0A0C9QW93 A0A3B0JWU7 A0A1A9X589 B0WQR9 A0A182N0N9 A0A1Q3FD85 Q29JE9 T1IAP9 A0A0A9X7R6 B4PZ50 B3NYF5 A0A0A9X6S5 A0A0K8TJ80 B4M886 A0A182QSP4 A0A0K8T7J7 A0A126CR36 A0A1W4V862 A0A182URY2 A0A226ERG3 A0A1B6JRL3 A0A182M5G9 B4L1Z6 A0A0K8V6T5 A0A182WZ94 A0A182XYB2 Q7KMM4 B4JNR8 A0A182R253 A0A182PUA8 Q7Q4V0 A0A1A9UUR7 A0A182HMR5 W8C608 A0A182VVG0 A0A0M4F9S7 A0A3P9B7H3 A0A182TVW1 A0A3B5A1R0 A0A2M3YZB8 A0A2H8TSQ9 A0A1B0G4M6 A0A3Q2HNI5 J9K948 A0A2S2NLM4 A0A1B0AX59 A0A3Q2ZSP1
Pubmed
23376632
19121390
26354079
22118469
29258441
18362917
+ More
19820115 24845553 24508170 30249741 20798317 28004739 21719571 21347285 21282665 17510324 27414796 17994087 25315136 28648823 24438588 26108605 15632085 23185243 25401762 26823975 17550304 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 24495485 25186727 19892987
19820115 24845553 24508170 30249741 20798317 28004739 21719571 21347285 21282665 17510324 27414796 17994087 25315136 28648823 24438588 26108605 15632085 23185243 25401762 26823975 17550304 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 24495485 25186727 19892987
EMBL
AB749778
BAM78681.1
BABH01024200
AB747260
BAM78680.1
KQ458995
+ More
KPJ04412.1 AGBW02009925 OWR49561.1 MH071438 AXJ21599.1 ODYU01003875 SOQ43180.1 KQ971321 KYB28709.1 KZ288203 PBC33374.1 NEVH01001358 PNF42738.1 KQ434977 KZC12863.1 KK852829 KDR15365.1 KQ435793 KOX74221.1 KK107200 EZA55504.1 QOIP01000002 RLU25319.1 GANO01001734 JAB58137.1 KQ414617 KOC68138.1 UFQS01000048 UFQT01000048 SSW98616.1 SSX19002.1 GFDF01005263 JAV08821.1 GFDF01005262 JAV08822.1 GFDF01005265 JAV08819.1 GL448512 EFN84496.1 GFDF01005266 JAV08818.1 GFDF01005264 JAV08820.1 GFDF01005261 JAV08823.1 KQ981820 KYN35164.1 GEZM01057065 JAV72464.1 CVRI01000005 CRK87777.1 GL887596 EGI70745.1 KQ981082 KYN09690.1 ADTU01012699 KQ976424 KYM88291.1 KQ982254 KYQ58762.1 GL765847 EFZ15875.1 CH477674 EAT37392.1 GALX01002848 JAB65618.1 AJWK01011154 AJWK01011155 KU764421 AQS60668.1 GECZ01005779 JAS63990.1 KY618821 AQW43007.1 GL435626 EFN72883.1 CH964239 EDW81853.1 KA647615 AFP62244.1 NNAY01001254 OXU24606.1 ATLV01023343 KE525342 KFB48949.1 GFTR01008281 JAW08145.1 JRES01001480 KNC22571.1 KY031136 ATU82887.1 GBYB01008008 GBYB01008009 GBYB01008010 JAG77775.1 JAG77776.1 JAG77777.1 OUUW01000003 SPP77826.1 DS232044 EDS32993.1 GFDL01009509 JAV25536.1 CH379063 EAL32352.2 KRT05891.1 KRT05892.1 ACPB03006700 GBHO01027938 GDHC01013673 JAG15666.1 JAQ04956.1 CM000162 EDX03111.1 KRK07120.1 CH954180 EDV47634.1 GBHO01027940 GDHC01005211 JAG15664.1 JAQ13418.1 GBRD01000173 JAG65648.1 CH940653 EDW62362.2 AXCN02002234 GBRD01004328 JAG61493.1 KC702772 AHG54244.1 LNIX01000002 OXA60099.1 GECU01005954 JAT01753.1 AXCM01001137 CH933810 EDW07717.1 GDHF01032699 GDHF01017667 JAI19615.1 JAI34647.1 AF145625 AE014298 AAD38600.1 AAF45432.1 AAG22460.1 AAN08995.1 AAN08996.1 AAN08997.1 CH916371 EDV92361.1 AAAB01008963 EAA12063.4 APCN01004862 GAMC01004574 JAC01982.1 CP012528 ALC49045.1 GGFM01000868 MBW21619.1 GFXV01004433 MBW16238.1 CCAG010000522 ABLF02036812 GGMR01005409 MBY18028.1 JXJN01005087
KPJ04412.1 AGBW02009925 OWR49561.1 MH071438 AXJ21599.1 ODYU01003875 SOQ43180.1 KQ971321 KYB28709.1 KZ288203 PBC33374.1 NEVH01001358 PNF42738.1 KQ434977 KZC12863.1 KK852829 KDR15365.1 KQ435793 KOX74221.1 KK107200 EZA55504.1 QOIP01000002 RLU25319.1 GANO01001734 JAB58137.1 KQ414617 KOC68138.1 UFQS01000048 UFQT01000048 SSW98616.1 SSX19002.1 GFDF01005263 JAV08821.1 GFDF01005262 JAV08822.1 GFDF01005265 JAV08819.1 GL448512 EFN84496.1 GFDF01005266 JAV08818.1 GFDF01005264 JAV08820.1 GFDF01005261 JAV08823.1 KQ981820 KYN35164.1 GEZM01057065 JAV72464.1 CVRI01000005 CRK87777.1 GL887596 EGI70745.1 KQ981082 KYN09690.1 ADTU01012699 KQ976424 KYM88291.1 KQ982254 KYQ58762.1 GL765847 EFZ15875.1 CH477674 EAT37392.1 GALX01002848 JAB65618.1 AJWK01011154 AJWK01011155 KU764421 AQS60668.1 GECZ01005779 JAS63990.1 KY618821 AQW43007.1 GL435626 EFN72883.1 CH964239 EDW81853.1 KA647615 AFP62244.1 NNAY01001254 OXU24606.1 ATLV01023343 KE525342 KFB48949.1 GFTR01008281 JAW08145.1 JRES01001480 KNC22571.1 KY031136 ATU82887.1 GBYB01008008 GBYB01008009 GBYB01008010 JAG77775.1 JAG77776.1 JAG77777.1 OUUW01000003 SPP77826.1 DS232044 EDS32993.1 GFDL01009509 JAV25536.1 CH379063 EAL32352.2 KRT05891.1 KRT05892.1 ACPB03006700 GBHO01027938 GDHC01013673 JAG15666.1 JAQ04956.1 CM000162 EDX03111.1 KRK07120.1 CH954180 EDV47634.1 GBHO01027940 GDHC01005211 JAG15664.1 JAQ13418.1 GBRD01000173 JAG65648.1 CH940653 EDW62362.2 AXCN02002234 GBRD01004328 JAG61493.1 KC702772 AHG54244.1 LNIX01000002 OXA60099.1 GECU01005954 JAT01753.1 AXCM01001137 CH933810 EDW07717.1 GDHF01032699 GDHF01017667 JAI19615.1 JAI34647.1 AF145625 AE014298 AAD38600.1 AAF45432.1 AAG22460.1 AAN08995.1 AAN08996.1 AAN08997.1 CH916371 EDV92361.1 AAAB01008963 EAA12063.4 APCN01004862 GAMC01004574 JAC01982.1 CP012528 ALC49045.1 GGFM01000868 MBW21619.1 GFXV01004433 MBW16238.1 CCAG010000522 ABLF02036812 GGMR01005409 MBY18028.1 JXJN01005087
Proteomes
UP000005204
UP000053268
UP000007151
UP000007266
UP000242457
UP000005203
+ More
UP000235965 UP000076502 UP000027135 UP000053105 UP000053097 UP000279307 UP000053825 UP000008237 UP000078541 UP000183832 UP000007755 UP000078492 UP000005205 UP000078540 UP000075809 UP000075880 UP000008820 UP000092461 UP000000311 UP000095300 UP000007798 UP000095301 UP000215335 UP000030765 UP000037069 UP000268350 UP000091820 UP000002320 UP000075884 UP000001819 UP000015103 UP000002282 UP000008711 UP000008792 UP000075886 UP000192221 UP000075903 UP000198287 UP000075883 UP000009192 UP000076407 UP000076408 UP000000803 UP000001070 UP000075900 UP000075885 UP000007062 UP000078200 UP000075840 UP000075920 UP000092553 UP000265160 UP000075902 UP000261400 UP000092444 UP000002281 UP000007819 UP000092460 UP000264800
UP000235965 UP000076502 UP000027135 UP000053105 UP000053097 UP000279307 UP000053825 UP000008237 UP000078541 UP000183832 UP000007755 UP000078492 UP000005205 UP000078540 UP000075809 UP000075880 UP000008820 UP000092461 UP000000311 UP000095300 UP000007798 UP000095301 UP000215335 UP000030765 UP000037069 UP000268350 UP000091820 UP000002320 UP000075884 UP000001819 UP000015103 UP000002282 UP000008711 UP000008792 UP000075886 UP000192221 UP000075903 UP000198287 UP000075883 UP000009192 UP000076407 UP000076408 UP000000803 UP000001070 UP000075900 UP000075885 UP000007062 UP000078200 UP000075840 UP000075920 UP000092553 UP000265160 UP000075902 UP000261400 UP000092444 UP000002281 UP000007819 UP000092460 UP000264800
PRIDE
Interpro
Gene 3D
ProteinModelPortal
L8B8U1
H9ISA5
L8B8V1
A0A0N0PA04
A0A212F746
A0A345W861
+ More
A0A2H1VQT4 A0A139WL23 A0A2A3EPD6 A0A088A7S9 A0A2J7RPJ2 A0A154PM08 A0A067QY16 A0A0N0U565 A0A026WK76 A0A3L8DZF9 U5EKN8 A0A0L7RBC3 A0A336LLR5 A0A1L8DQZ3 A0A1L8DQT5 A0A1L8DQW6 E2BII6 A0A1L8DQR3 A0A1L8DR57 A0A1L8DQT4 A0A195F510 A0A1Y1LFQ8 A0A1J1HNH3 F4W549 A0A195D9U3 A0A158NDK9 A0A195BQQ2 A0A151XEG8 A0A182JCA9 E9IU82 Q16SF9 V5GYU2 A0A1B0ETK8 A0A1S6J0X3 A0A1B6GNL0 A0A2K8FTL6 E2A0I0 A0A1I8P7N9 B4NDN1 T1PHL5 A0A1I8MLM4 A0A232F1E1 A0A084WFF5 A0A224X6Q8 A0A0L0BRC7 A0A2K8JWU1 A0A0C9QW93 A0A3B0JWU7 A0A1A9X589 B0WQR9 A0A182N0N9 A0A1Q3FD85 Q29JE9 T1IAP9 A0A0A9X7R6 B4PZ50 B3NYF5 A0A0A9X6S5 A0A0K8TJ80 B4M886 A0A182QSP4 A0A0K8T7J7 A0A126CR36 A0A1W4V862 A0A182URY2 A0A226ERG3 A0A1B6JRL3 A0A182M5G9 B4L1Z6 A0A0K8V6T5 A0A182WZ94 A0A182XYB2 Q7KMM4 B4JNR8 A0A182R253 A0A182PUA8 Q7Q4V0 A0A1A9UUR7 A0A182HMR5 W8C608 A0A182VVG0 A0A0M4F9S7 A0A3P9B7H3 A0A182TVW1 A0A3B5A1R0 A0A2M3YZB8 A0A2H8TSQ9 A0A1B0G4M6 A0A3Q2HNI5 J9K948 A0A2S2NLM4 A0A1B0AX59 A0A3Q2ZSP1
A0A2H1VQT4 A0A139WL23 A0A2A3EPD6 A0A088A7S9 A0A2J7RPJ2 A0A154PM08 A0A067QY16 A0A0N0U565 A0A026WK76 A0A3L8DZF9 U5EKN8 A0A0L7RBC3 A0A336LLR5 A0A1L8DQZ3 A0A1L8DQT5 A0A1L8DQW6 E2BII6 A0A1L8DQR3 A0A1L8DR57 A0A1L8DQT4 A0A195F510 A0A1Y1LFQ8 A0A1J1HNH3 F4W549 A0A195D9U3 A0A158NDK9 A0A195BQQ2 A0A151XEG8 A0A182JCA9 E9IU82 Q16SF9 V5GYU2 A0A1B0ETK8 A0A1S6J0X3 A0A1B6GNL0 A0A2K8FTL6 E2A0I0 A0A1I8P7N9 B4NDN1 T1PHL5 A0A1I8MLM4 A0A232F1E1 A0A084WFF5 A0A224X6Q8 A0A0L0BRC7 A0A2K8JWU1 A0A0C9QW93 A0A3B0JWU7 A0A1A9X589 B0WQR9 A0A182N0N9 A0A1Q3FD85 Q29JE9 T1IAP9 A0A0A9X7R6 B4PZ50 B3NYF5 A0A0A9X6S5 A0A0K8TJ80 B4M886 A0A182QSP4 A0A0K8T7J7 A0A126CR36 A0A1W4V862 A0A182URY2 A0A226ERG3 A0A1B6JRL3 A0A182M5G9 B4L1Z6 A0A0K8V6T5 A0A182WZ94 A0A182XYB2 Q7KMM4 B4JNR8 A0A182R253 A0A182PUA8 Q7Q4V0 A0A1A9UUR7 A0A182HMR5 W8C608 A0A182VVG0 A0A0M4F9S7 A0A3P9B7H3 A0A182TVW1 A0A3B5A1R0 A0A2M3YZB8 A0A2H8TSQ9 A0A1B0G4M6 A0A3Q2HNI5 J9K948 A0A2S2NLM4 A0A1B0AX59 A0A3Q2ZSP1
PDB
5HJR
E-value=0,
Score=2255
Ontologies
PATHWAY
GO
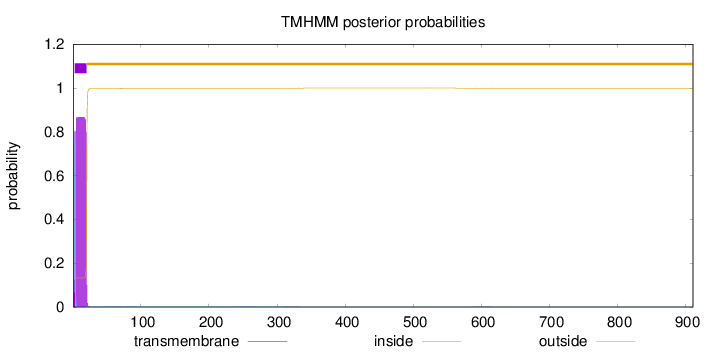
Topology
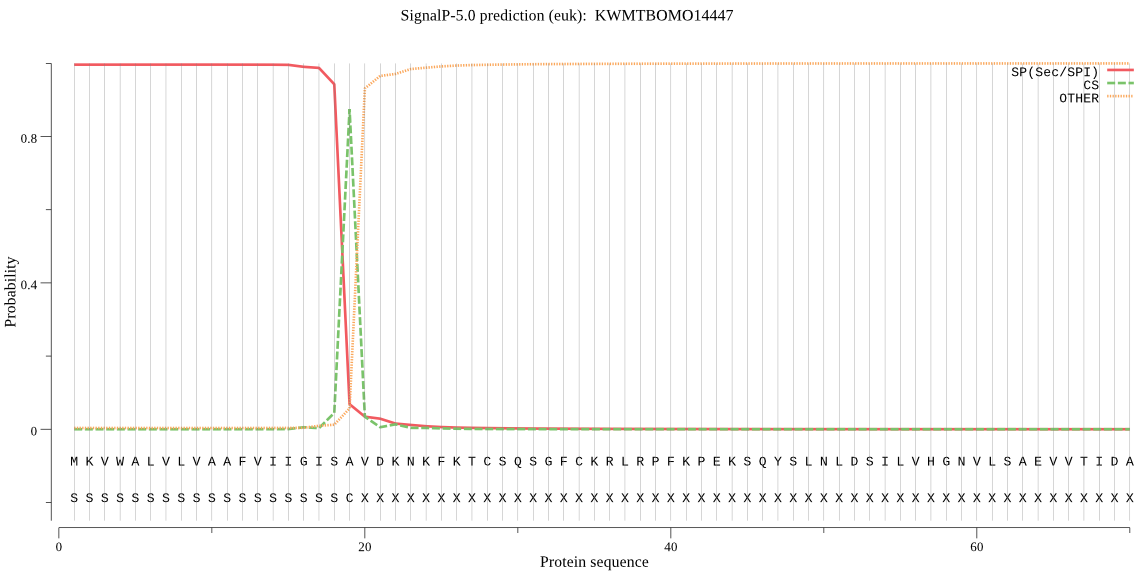
SignalP
Position: 1 - 19,
Likelihood: 0.996045
Length:
911
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.68985
Exp number, first 60 AAs:
15.64928
Total prob of N-in:
0.86558
POSSIBLE N-term signal
sequence
inside
1 - 2
TMhelix
3 - 20
outside
21 - 911
Population Genetic Test Statistics
Pi
230.567827
Theta
176.030597
Tajima's D
1.478727
CLR
0.033987
CSRT
0.785510724463777
Interpretation
Uncertain
