Gene
KWMTBOMO14446 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000086
Annotation
LRP16_protein_[Bombyx_mori]
Full name
ADP-ribose glycohydrolase MACROD2
Alternative Name
MACRO domain-containing protein 2
O-acetyl-ADP-ribose deacetylase MACROD2
[Protein ADP-ribosylaspartate] hydrolase MACROD2
[Protein ADP-ribosylglutamate] hydrolase MACROD2
O-acetyl-ADP-ribose deacetylase MACROD2
[Protein ADP-ribosylaspartate] hydrolase MACROD2
[Protein ADP-ribosylglutamate] hydrolase MACROD2
Location in the cell
Cytoplasmic Reliability : 1.754 Mitochondrial Reliability : 1.077 Nuclear Reliability : 1.141
Sequence
CDS
ATGGTTAATTCTACTAAGTGGGAAATAGAAAAGAACAGGATTCTTAAACTTTCATTAGAAGAGAAGAGAAAAATCTATAAATCTTCTGATTTCATAGATTTAGAGAATGTTGATCCTTGGAGTAAATATCTTAACAAAAGTCAAGGAATTGATTCCAAGAAAAGCACCACAGACGATTTGAAGGAGTTTGAGAAGATAAAGATAAACACAGAGAAGAATAAATCCATTTCTGAGCGCGTTTCTATTTTTAAAGGAGACATAACTAAGCTGGAAATTGATGCAGTTGTGAATGCTGCTAATTCAAGACTAAAGGCTGGAGGTGGTGTCGATGGAGCCATACACCGGGCTGCAGGTCCCTTTTTACAGGCAGAATGCGATTCTATAGGCGGATGTCCAACAGGAGATGCCAAAGTTACTGGTGGTTACAATTTACCAGCTAAATATATCATCCACACTGTTGGGCCCCAAGATGGATCGGCAGAGAAACTAGAATCCTGCTACGAAAAATGCCTGTCATTCCAACAGGAATACCAAATAAAATCGATCGCTTTTCCGTGTATATCCACTGGAATTTACGGTTTTCCGAATCGTCTCGCAGCACACATAGCGCTACGTACAGCTAGGAAGTTTCTAGAAACGAATACAGAAATGAACCGAATTATATTTTGCACGTTCTTACCTATCGATGTGGAGATTTACGAGACGTTGATGCAACTATACTTTCCTACACTTTAA
Protein
MVNSTKWEIEKNRILKLSLEEKRKIYKSSDFIDLENVDPWSKYLNKSQGIDSKKSTTDDLKEFEKIKINTEKNKSISERVSIFKGDITKLEIDAVVNAANSRLKAGGGVDGAIHRAAGPFLQAECDSIGGCPTGDAKVTGGYNLPAKYIIHTVGPQDGSAEKLESCYEKCLSFQQEYQIKSIAFPCISTGIYGFPNRLAAHIALRTARKFLETNTEMNRIIFCTFLPIDVEIYETLMQLYFPTL
Summary
Description
Removes ADP-ribose from asparatate and glutamate residues in proteins bearing a single ADP-ribose moiety. Inactive towards proteins bearing poly-ADP-ribose. Deacetylates O-acetyl-ADP ribose, a signaling molecule generated by the deacetylation of acetylated lysine residues in histones and other proteins.
Catalytic Activity
2''-O-acetyl-ADP-D-ribose + H2O = acetate + ADP-D-ribose + H(+)
4-O-(ADP-D-ribosyl)-L-aspartyl-[protein] + H2O = ADP-D-ribose + H(+) + L-aspartyl-[protein]
5-O-(ADP-D-ribosyl)-L-glutamyl-[protein] + H2O = ADP-D-ribose + H(+) + L-glutamyl-[protein]
4-O-(ADP-D-ribosyl)-L-aspartyl-[protein] + H2O = ADP-D-ribose + H(+) + L-aspartyl-[protein]
5-O-(ADP-D-ribosyl)-L-glutamyl-[protein] + H2O = ADP-D-ribose + H(+) + L-glutamyl-[protein]
Subunit
Interacts with ADP-ribosylated PARP1.
Keywords
Complete proteome
DNA damage
Hydrolase
Isopeptide bond
Nucleus
Reference proteome
Ubl conjugation
Feature
chain ADP-ribose glycohydrolase MACROD2
Uniprot
H9IS60
Q1HPZ5
S4P8Y2
A0A0N1IHF9
A0A0N1PHE1
A0A212F743
+ More
A0A2H1VS96 A0A2A4K9G0 A0A2W1BLD8 A0A2L2Y8W3 A0A087T7L1 A0A067QYM6 D6X1V2 E2AVU0 A0A226E2B3 R4WJN2 V9IDA7 A0A0J7LAB6 A0A131ZD45 A0A131XPL2 A0A146LEE7 A0A2A3EP00 A0A088AU96 A0A131Z4N3 F4W7Z0 A0A224Z3A3 A0A1E1XBL8 A0A0A9VSM3 A0A195EZC4 A0A1B6CL41 E2BKS9 A0A195D7T8 A0A1B6F8Y7 A0A0A9YWS5 A0A0A9VSM6 A0A1B6FER4 A0A1B6F0H1 A0A3L8DFK8 A0A0A9YV72 A0A026WR31 A0A1Z5L2I7 B7PF53 A0A1B6JZ62 A0A131Y1D1 A0A1S3JK74 A0A195C534 A0A1B6IAR3 A0A1B6MFK4 G3MI23 A0A1B6LMS9 N6TNC6 A0A2D4MGF9 A0A158NDU1 U4UYG7 A0A0V0G4D9 J3JWP8 A0A0P4VLG1 R4G5Y2 A0A1S3JLI6 A0A1Y1MS46 A0A151X471 A0A154PG99 A0A2J7Q4B6 A0A218URH4 A0A2J7Q4D0 A0A2C9K3W2 A0A091J3H5 A0A293LAS1 A0A2Y9QV05 A0A195BQM5 K7ISE1 A0A2R5LCE7 A2AS33 A0A2J7Q4B5 A0A3B4GDL3 A0A3B4GC55 A0A3P9B7V4 A0A3Q2VJ10 A0A3Q2WQA1 A0A1B6JN87 A0A232EN17 G3PXL1 A0A166BW11 A0A2D0RID8 A0A2D0RID7 Q3UYG8 A0A3B4UY62 A0A2Y9HXW7 A0A2Y9IRG6 A0A3P9MJC0 A0A0P6GIQ1 A0A0P5QIS9 A0A3P9I856 A0A2Y9PSM4 A0A162P386 A0A3Q7SNK7 A0A3B4BUI3 A0A3B4XX66
A0A2H1VS96 A0A2A4K9G0 A0A2W1BLD8 A0A2L2Y8W3 A0A087T7L1 A0A067QYM6 D6X1V2 E2AVU0 A0A226E2B3 R4WJN2 V9IDA7 A0A0J7LAB6 A0A131ZD45 A0A131XPL2 A0A146LEE7 A0A2A3EP00 A0A088AU96 A0A131Z4N3 F4W7Z0 A0A224Z3A3 A0A1E1XBL8 A0A0A9VSM3 A0A195EZC4 A0A1B6CL41 E2BKS9 A0A195D7T8 A0A1B6F8Y7 A0A0A9YWS5 A0A0A9VSM6 A0A1B6FER4 A0A1B6F0H1 A0A3L8DFK8 A0A0A9YV72 A0A026WR31 A0A1Z5L2I7 B7PF53 A0A1B6JZ62 A0A131Y1D1 A0A1S3JK74 A0A195C534 A0A1B6IAR3 A0A1B6MFK4 G3MI23 A0A1B6LMS9 N6TNC6 A0A2D4MGF9 A0A158NDU1 U4UYG7 A0A0V0G4D9 J3JWP8 A0A0P4VLG1 R4G5Y2 A0A1S3JLI6 A0A1Y1MS46 A0A151X471 A0A154PG99 A0A2J7Q4B6 A0A218URH4 A0A2J7Q4D0 A0A2C9K3W2 A0A091J3H5 A0A293LAS1 A0A2Y9QV05 A0A195BQM5 K7ISE1 A0A2R5LCE7 A2AS33 A0A2J7Q4B5 A0A3B4GDL3 A0A3B4GC55 A0A3P9B7V4 A0A3Q2VJ10 A0A3Q2WQA1 A0A1B6JN87 A0A232EN17 G3PXL1 A0A166BW11 A0A2D0RID8 A0A2D0RID7 Q3UYG8 A0A3B4UY62 A0A2Y9HXW7 A0A2Y9IRG6 A0A3P9MJC0 A0A0P6GIQ1 A0A0P5QIS9 A0A3P9I856 A0A2Y9PSM4 A0A162P386 A0A3Q7SNK7 A0A3B4BUI3 A0A3B4XX66
Pubmed
19121390
23622113
26354079
22118469
28756777
26561354
+ More
24845553 18362917 19820115 20798317 23691247 26830274 28049606 26823975 21719571 28797301 28503490 25401762 30249741 24508170 28528879 22216098 23537049 21347285 22516182 27129103 28004739 15562597 20075255 19468303 21183079 25186727 28648823 26659563 16141072 17586838 17554307
24845553 18362917 19820115 20798317 23691247 26830274 28049606 26823975 21719571 28797301 28503490 25401762 30249741 24508170 28528879 22216098 23537049 21347285 22516182 27129103 28004739 15562597 20075255 19468303 21183079 25186727 28648823 26659563 16141072 17586838 17554307
EMBL
BABH01024199
DQ443257
ABF51346.1
GAIX01003979
JAA88581.1
KQ460195
+ More
KPJ17208.1 KQ458995 KPJ04411.1 AGBW02009925 OWR49562.1 ODYU01004135 SOQ43689.1 NWSH01000010 PCG80925.1 KZ150182 PZC72493.1 IAAA01011206 IAAA01011207 IAAA01011208 LAA04619.1 KK113815 KFM61100.1 KK852820 KDR15591.1 KQ971371 EFA09957.2 GL443213 EFN62362.1 LNIX01000007 OXA51703.1 AK417780 BAN20995.1 JR039304 AEY58627.1 LBMM01000059 KMR05131.1 GEDV01000141 JAP88416.1 GEFH01000426 JAP68155.1 GDHC01012041 JAQ06588.1 KZ288203 PBC33447.1 GEDV01002619 JAP85938.1 GL887888 EGI69660.1 GFPF01009548 MAA20694.1 GFAC01002589 JAT96599.1 GBHO01044880 JAF98723.1 KQ981906 KYN33224.1 GEDC01023185 GEDC01000019 JAS14113.1 JAS37279.1 GL448826 EFN83678.1 KQ981153 KYN08928.1 GECZ01023158 JAS46611.1 GBHO01007533 JAG36071.1 GBHO01044875 GBHO01007534 GBHO01007527 JAF98728.1 JAG36070.1 JAG36077.1 GECZ01021074 GECZ01009389 JAS48695.1 JAS60380.1 GECZ01026071 JAS43698.1 QOIP01000008 RLU19250.1 GBHO01044888 GBHO01007530 JAF98715.1 JAG36074.1 KK107126 EZA58433.1 GFJQ02005739 JAW01231.1 ABJB010183252 ABJB010520409 DS699372 EEC05225.1 GECU01003225 JAT04482.1 GEFM01003960 JAP71836.1 KQ978251 KYM95977.1 GECU01023698 JAS84008.1 GEBQ01008392 GEBQ01005302 JAT31585.1 JAT34675.1 JO841524 AEO33141.1 GEBQ01015010 JAT24967.1 APGK01058003 APGK01058004 APGK01058005 KB741285 ENN70745.1 IACM01100255 LAB32464.1 ADTU01012982 KB632425 ERL95355.1 GECL01003225 JAP02899.1 BT127666 AEE62628.1 GDKW01003109 JAI53486.1 GAHY01000120 JAA77390.1 GEZM01027147 JAV86816.1 KQ982548 KYQ55235.1 KQ434899 KZC10906.1 NEVH01018385 PNF23426.1 MUZQ01000159 OWK56407.1 PNF23425.1 KL218527 KFP06336.1 GFWV01000128 MAA24858.1 KQ976424 KYM88438.1 GGLE01002901 MBY07027.1 AL731795 AL837516 AL844562 AL844582 AL928700 AL929027 AL929380 AL953906 BX294658 BX296531 PNF23424.1 GECU01007002 JAT00705.1 NNAY01003253 OXU19728.1 KV424619 KZV65532.1 AK134694 AL845530 AL928823 AL929443 GDIQ01032683 JAN62054.1 GDIQ01134590 JAL17136.1 LRGB01000512 KZS18480.1
KPJ17208.1 KQ458995 KPJ04411.1 AGBW02009925 OWR49562.1 ODYU01004135 SOQ43689.1 NWSH01000010 PCG80925.1 KZ150182 PZC72493.1 IAAA01011206 IAAA01011207 IAAA01011208 LAA04619.1 KK113815 KFM61100.1 KK852820 KDR15591.1 KQ971371 EFA09957.2 GL443213 EFN62362.1 LNIX01000007 OXA51703.1 AK417780 BAN20995.1 JR039304 AEY58627.1 LBMM01000059 KMR05131.1 GEDV01000141 JAP88416.1 GEFH01000426 JAP68155.1 GDHC01012041 JAQ06588.1 KZ288203 PBC33447.1 GEDV01002619 JAP85938.1 GL887888 EGI69660.1 GFPF01009548 MAA20694.1 GFAC01002589 JAT96599.1 GBHO01044880 JAF98723.1 KQ981906 KYN33224.1 GEDC01023185 GEDC01000019 JAS14113.1 JAS37279.1 GL448826 EFN83678.1 KQ981153 KYN08928.1 GECZ01023158 JAS46611.1 GBHO01007533 JAG36071.1 GBHO01044875 GBHO01007534 GBHO01007527 JAF98728.1 JAG36070.1 JAG36077.1 GECZ01021074 GECZ01009389 JAS48695.1 JAS60380.1 GECZ01026071 JAS43698.1 QOIP01000008 RLU19250.1 GBHO01044888 GBHO01007530 JAF98715.1 JAG36074.1 KK107126 EZA58433.1 GFJQ02005739 JAW01231.1 ABJB010183252 ABJB010520409 DS699372 EEC05225.1 GECU01003225 JAT04482.1 GEFM01003960 JAP71836.1 KQ978251 KYM95977.1 GECU01023698 JAS84008.1 GEBQ01008392 GEBQ01005302 JAT31585.1 JAT34675.1 JO841524 AEO33141.1 GEBQ01015010 JAT24967.1 APGK01058003 APGK01058004 APGK01058005 KB741285 ENN70745.1 IACM01100255 LAB32464.1 ADTU01012982 KB632425 ERL95355.1 GECL01003225 JAP02899.1 BT127666 AEE62628.1 GDKW01003109 JAI53486.1 GAHY01000120 JAA77390.1 GEZM01027147 JAV86816.1 KQ982548 KYQ55235.1 KQ434899 KZC10906.1 NEVH01018385 PNF23426.1 MUZQ01000159 OWK56407.1 PNF23425.1 KL218527 KFP06336.1 GFWV01000128 MAA24858.1 KQ976424 KYM88438.1 GGLE01002901 MBY07027.1 AL731795 AL837516 AL844562 AL844582 AL928700 AL929027 AL929380 AL953906 BX294658 BX296531 PNF23424.1 GECU01007002 JAT00705.1 NNAY01003253 OXU19728.1 KV424619 KZV65532.1 AK134694 AL845530 AL928823 AL929443 GDIQ01032683 JAN62054.1 GDIQ01134590 JAL17136.1 LRGB01000512 KZS18480.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000054359
+ More
UP000027135 UP000007266 UP000000311 UP000198287 UP000036403 UP000242457 UP000005203 UP000007755 UP000078541 UP000008237 UP000078492 UP000279307 UP000053097 UP000001555 UP000085678 UP000078542 UP000019118 UP000005205 UP000030742 UP000075809 UP000076502 UP000235965 UP000197619 UP000076420 UP000054308 UP000248480 UP000078540 UP000002358 UP000000589 UP000261460 UP000265160 UP000264840 UP000215335 UP000007635 UP000077086 UP000221080 UP000261420 UP000248481 UP000248482 UP000265180 UP000265200 UP000248483 UP000076858 UP000286640 UP000261440 UP000261360
UP000027135 UP000007266 UP000000311 UP000198287 UP000036403 UP000242457 UP000005203 UP000007755 UP000078541 UP000008237 UP000078492 UP000279307 UP000053097 UP000001555 UP000085678 UP000078542 UP000019118 UP000005205 UP000030742 UP000075809 UP000076502 UP000235965 UP000197619 UP000076420 UP000054308 UP000248480 UP000078540 UP000002358 UP000000589 UP000261460 UP000265160 UP000264840 UP000215335 UP000007635 UP000077086 UP000221080 UP000261420 UP000248481 UP000248482 UP000265180 UP000265200 UP000248483 UP000076858 UP000286640 UP000261440 UP000261360
PRIDE
Pfam
PF01661 Macro
Interpro
IPR002589
Macro_dom
ProteinModelPortal
H9IS60
Q1HPZ5
S4P8Y2
A0A0N1IHF9
A0A0N1PHE1
A0A212F743
+ More
A0A2H1VS96 A0A2A4K9G0 A0A2W1BLD8 A0A2L2Y8W3 A0A087T7L1 A0A067QYM6 D6X1V2 E2AVU0 A0A226E2B3 R4WJN2 V9IDA7 A0A0J7LAB6 A0A131ZD45 A0A131XPL2 A0A146LEE7 A0A2A3EP00 A0A088AU96 A0A131Z4N3 F4W7Z0 A0A224Z3A3 A0A1E1XBL8 A0A0A9VSM3 A0A195EZC4 A0A1B6CL41 E2BKS9 A0A195D7T8 A0A1B6F8Y7 A0A0A9YWS5 A0A0A9VSM6 A0A1B6FER4 A0A1B6F0H1 A0A3L8DFK8 A0A0A9YV72 A0A026WR31 A0A1Z5L2I7 B7PF53 A0A1B6JZ62 A0A131Y1D1 A0A1S3JK74 A0A195C534 A0A1B6IAR3 A0A1B6MFK4 G3MI23 A0A1B6LMS9 N6TNC6 A0A2D4MGF9 A0A158NDU1 U4UYG7 A0A0V0G4D9 J3JWP8 A0A0P4VLG1 R4G5Y2 A0A1S3JLI6 A0A1Y1MS46 A0A151X471 A0A154PG99 A0A2J7Q4B6 A0A218URH4 A0A2J7Q4D0 A0A2C9K3W2 A0A091J3H5 A0A293LAS1 A0A2Y9QV05 A0A195BQM5 K7ISE1 A0A2R5LCE7 A2AS33 A0A2J7Q4B5 A0A3B4GDL3 A0A3B4GC55 A0A3P9B7V4 A0A3Q2VJ10 A0A3Q2WQA1 A0A1B6JN87 A0A232EN17 G3PXL1 A0A166BW11 A0A2D0RID8 A0A2D0RID7 Q3UYG8 A0A3B4UY62 A0A2Y9HXW7 A0A2Y9IRG6 A0A3P9MJC0 A0A0P6GIQ1 A0A0P5QIS9 A0A3P9I856 A0A2Y9PSM4 A0A162P386 A0A3Q7SNK7 A0A3B4BUI3 A0A3B4XX66
A0A2H1VS96 A0A2A4K9G0 A0A2W1BLD8 A0A2L2Y8W3 A0A087T7L1 A0A067QYM6 D6X1V2 E2AVU0 A0A226E2B3 R4WJN2 V9IDA7 A0A0J7LAB6 A0A131ZD45 A0A131XPL2 A0A146LEE7 A0A2A3EP00 A0A088AU96 A0A131Z4N3 F4W7Z0 A0A224Z3A3 A0A1E1XBL8 A0A0A9VSM3 A0A195EZC4 A0A1B6CL41 E2BKS9 A0A195D7T8 A0A1B6F8Y7 A0A0A9YWS5 A0A0A9VSM6 A0A1B6FER4 A0A1B6F0H1 A0A3L8DFK8 A0A0A9YV72 A0A026WR31 A0A1Z5L2I7 B7PF53 A0A1B6JZ62 A0A131Y1D1 A0A1S3JK74 A0A195C534 A0A1B6IAR3 A0A1B6MFK4 G3MI23 A0A1B6LMS9 N6TNC6 A0A2D4MGF9 A0A158NDU1 U4UYG7 A0A0V0G4D9 J3JWP8 A0A0P4VLG1 R4G5Y2 A0A1S3JLI6 A0A1Y1MS46 A0A151X471 A0A154PG99 A0A2J7Q4B6 A0A218URH4 A0A2J7Q4D0 A0A2C9K3W2 A0A091J3H5 A0A293LAS1 A0A2Y9QV05 A0A195BQM5 K7ISE1 A0A2R5LCE7 A2AS33 A0A2J7Q4B5 A0A3B4GDL3 A0A3B4GC55 A0A3P9B7V4 A0A3Q2VJ10 A0A3Q2WQA1 A0A1B6JN87 A0A232EN17 G3PXL1 A0A166BW11 A0A2D0RID8 A0A2D0RID7 Q3UYG8 A0A3B4UY62 A0A2Y9HXW7 A0A2Y9IRG6 A0A3P9MJC0 A0A0P6GIQ1 A0A0P5QIS9 A0A3P9I856 A0A2Y9PSM4 A0A162P386 A0A3Q7SNK7 A0A3B4BUI3 A0A3B4XX66
PDB
4IQY
E-value=2.42728e-59,
Score=577
Ontologies
GO
Topology
Subcellular location
Nucleus
Recruited to DNA lesions, probably via mono-APD-ribosylated proteins. With evidence from 1 publications.
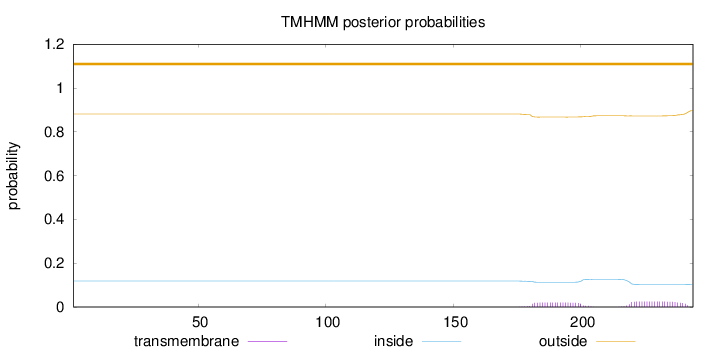
Length:
244
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.94879
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11855
outside
1 - 244
Population Genetic Test Statistics
Pi
185.036329
Theta
154.80693
Tajima's D
0.415669
CLR
337.28118
CSRT
0.490325483725814
Interpretation
Uncertain
