Gene
KWMTBOMO14443
Pre Gene Modal
BGIBMGA000088
Annotation
Seipin_[Papilio_xuthus]
Full name
Seipin
Location in the cell
Cytoplasmic Reliability : 1.659
Sequence
CDS
ATGCCTAATATAACACATCAGCGTACTGTACATTTTCAATTTAAGCCATGTGAAGAAACCATGGGGATATGTTCTTTTCCATATGCCTACGTGCAATTGACAAGAAAGAGCTCCATACTGATGTCAGGCCAGTTGTACCGCATAAGGCTAATTTTGGAAATGCCAGAGTCAAATGTCAACAAGGATCTCGGAATGTTCATGGTGTGCACACAGATGAGAGCAAAGGGTGGAGTTTTAGTGTCTTCATCATGTCGCTCAACTATGTTAAGATACAGGTCGAAACTCCACGAATACATCCGCACAGCGGTCATGGCGCCTCTCCTCATGTCGGACGTGGTGGAAGAGAAGCAGACCATCCAAGTCGATTTGTTCACGGAATTCGATGATGATCCCAATCAACCCGTTACGGACGCGTACGTCGAACTCCAGTCGCGCTACGTGCAAGTGTACGGAGGTCAGCTACACATCGAGGCTCACTTCACCGGCCTCCGTTACCTGATGTACAACTGGCCTAAGTTCTCGGCTCTCTTCGGCATCAGCACTAATCTGTTCTTCATAACGCTCGTGTTCGCGCTGAGCTGGTATCACCTACAGGAAGGGTTGCCAGAGTTTATCAAGAGTAAACTGGGTACGACGCCGGACAAAGATGAAGACAATAAGAAATTAATCGGAAAGATAAAATTAGAAAAAGAAGAGTCGCCTTCCATGTTTGAAGGCGACGTGCTTCTAGAAGAGTTTATGCAGCTCGAAGAGAAAGAGAAGAAGACATCGTAG
Protein
MPNITHQRTVHFQFKPCEETMGICSFPYAYVQLTRKSSILMSGQLYRIRLILEMPESNVNKDLGMFMVCTQMRAKGGVLVSSSCRSTMLRYRSKLHEYIRTAVMAPLLMSDVVEEKQTIQVDLFTEFDDDPNQPVTDAYVELQSRYVQVYGGQLHIEAHFTGLRYLMYNWPKFSALFGISTNLFFITLVFALSWYHLQEGLPEFIKSKLGTTPDKDEDNKKLIGKIKLEKEESPSMFEGDVLLEEFMQLEEKEKKTS
Summary
Description
Is a regulator of lipid catabolism essential for adipocyte differentiation. May also be involved in the central regulation of energy homeostasis (By similarity). Necessary for correct lipid storage and lipid droplets maintenance; plays a tissue-autonomous role in controlling lipid storage in adipocytes and in preventing ectopic lipid droplet formation in non-adipose tissues. May participate in phosphatidic acid metabolism and subsequently down-regulate lipogenesis.
Keywords
3D-structure
Complete proteome
Endoplasmic reticulum
Lipid degradation
Lipid metabolism
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Seipin
Uniprot
A0A2W1BH21
A0A0N1IAH1
A0A1E1WPI6
A0A2H1WAY8
A0A1E1WBA5
A0A2A4K9M2
+ More
A0A1B6KSS0 A0A1B6HVA4 A0A067R0Z7 A0A1B6GRF3 A0A2P8YC43 A0A088AE92 A0A2A3ET98 A0A2J7R455 A0A1W4XS61 A0A1S3D724 K7JF93 A0A232EG66 A0A023FA80 A0A1Y1LIE7 A0A1Y1LIG9 A0A151IMA1 E2C419 A0A0T6B7S2 A0A1D2NKC8 A0A026W2T2 A0A154P6K5 A0A0P4VPG3 A0A0K8TWF7 A0A1A9X425 A0A0M8ZYE8 A0A0K8WLI6 A0A336MJT5 A0A195FCK4 W8C413 A0A034WAD4 A0A0M4EW59 A0A1A9ZFY6 A0A1B0FBL8 E9IJH8 A0A1A9V2Z0 A0A0A1XFG9 B4JLF3 A0A1I8N2L7 B4M3F5 A0A1I8PSZ1 Q9V3X4 V5I9U7 A0A158NBT0 B4L426 A0A195BG91 A0A151J9A0 E0VDD6 A0A0A9YMV8 A0A146M715 A0A3B0K831 Q29IM5 A0A0A9YH79 A0A3B0K9V8 A0A146MDX3 A0A146LYD4 B4N1K8 A0A1W4VTQ9 B3MYZ6 A0A151X9F0 A0A0A9YMV3 F4X8B5 B4I9X3 B3P937 A0A2R7X289 R4WEE9 A0A0J7L074 A0A224XYT7 A0A1L8DJI7 A0A1L8DJV7 A0A1B6CYY4 J3JWT9 V5GWQ7 A0A226EU41 T1PKQ2 A0A1Q3FG76 B0WB41 A0A1Q3FFK4 A0A0L0C6W5 A0A182XVJ4 A0A084VZX9 A0A2S2QVC6 A0A182K872 E9FSR3 A0A182QB60 A0A182MWD0 A0A182J0P3 A0A182PTM7 A0A182S6R6 A0A1Y9J159 Q7QBJ6 A0A182HJ35 A0A182UKU8
A0A1B6KSS0 A0A1B6HVA4 A0A067R0Z7 A0A1B6GRF3 A0A2P8YC43 A0A088AE92 A0A2A3ET98 A0A2J7R455 A0A1W4XS61 A0A1S3D724 K7JF93 A0A232EG66 A0A023FA80 A0A1Y1LIE7 A0A1Y1LIG9 A0A151IMA1 E2C419 A0A0T6B7S2 A0A1D2NKC8 A0A026W2T2 A0A154P6K5 A0A0P4VPG3 A0A0K8TWF7 A0A1A9X425 A0A0M8ZYE8 A0A0K8WLI6 A0A336MJT5 A0A195FCK4 W8C413 A0A034WAD4 A0A0M4EW59 A0A1A9ZFY6 A0A1B0FBL8 E9IJH8 A0A1A9V2Z0 A0A0A1XFG9 B4JLF3 A0A1I8N2L7 B4M3F5 A0A1I8PSZ1 Q9V3X4 V5I9U7 A0A158NBT0 B4L426 A0A195BG91 A0A151J9A0 E0VDD6 A0A0A9YMV8 A0A146M715 A0A3B0K831 Q29IM5 A0A0A9YH79 A0A3B0K9V8 A0A146MDX3 A0A146LYD4 B4N1K8 A0A1W4VTQ9 B3MYZ6 A0A151X9F0 A0A0A9YMV3 F4X8B5 B4I9X3 B3P937 A0A2R7X289 R4WEE9 A0A0J7L074 A0A224XYT7 A0A1L8DJI7 A0A1L8DJV7 A0A1B6CYY4 J3JWT9 V5GWQ7 A0A226EU41 T1PKQ2 A0A1Q3FG76 B0WB41 A0A1Q3FFK4 A0A0L0C6W5 A0A182XVJ4 A0A084VZX9 A0A2S2QVC6 A0A182K872 E9FSR3 A0A182QB60 A0A182MWD0 A0A182J0P3 A0A182PTM7 A0A182S6R6 A0A1Y9J159 Q7QBJ6 A0A182HJ35 A0A182UKU8
Pubmed
28756777
26354079
24845553
29403074
20075255
28648823
+ More
25474469 28004739 20798317 27289101 24508170 30249741 27129103 24495485 25348373 21282665 25830018 17994087 25315136 10731132 12537572 10731137 12537569 21533227 21347285 20566863 25401762 26823975 15632085 21719571 23691247 22516182 23537049 26108605 25244985 24438588 21292972 12364791 14747013 17210077
25474469 28004739 20798317 27289101 24508170 30249741 27129103 24495485 25348373 21282665 25830018 17994087 25315136 10731132 12537572 10731137 12537569 21533227 21347285 20566863 25401762 26823975 15632085 21719571 23691247 22516182 23537049 26108605 25244985 24438588 21292972 12364791 14747013 17210077
EMBL
KZ150182
PZC72497.1
KQ458995
KPJ04408.1
GDQN01005789
GDQN01002129
+ More
GDQN01000295 JAT85265.1 JAT88925.1 JAT90759.1 ODYU01007447 SOQ50228.1 GDQN01006837 JAT84217.1 NWSH01000010 PCG80931.1 GEBQ01025475 JAT14502.1 GECU01029102 JAS78604.1 KK852820 KDR15578.1 GECZ01004758 JAS65011.1 PYGN01000712 PSN41829.1 KZ288192 PBC34369.1 NEVH01007815 PNF35614.1 AAZX01000596 NNAY01004846 OXU17346.1 GBBI01000330 JAC18382.1 GEZM01054748 JAV73424.1 GEZM01054749 JAV73423.1 KQ977059 KYN06018.1 GL452364 EFN77405.1 LJIG01009419 KRT83117.1 LJIJ01000018 ODN05679.1 KK107459 QOIP01000004 EZA50387.1 RLU23713.1 KQ434827 KZC07497.1 GDKW01003552 JAI53043.1 GDHF01033525 JAI18789.1 KQ435794 KOX73640.1 GDHF01010467 GDHF01000405 JAI41847.1 JAI51909.1 UFQS01000668 UFQT01000668 SSX05971.1 SSX26328.1 KQ981673 KYN38370.1 GAMC01002542 JAC04014.1 GAKP01007699 GAKP01007697 GAKP01007695 GAKP01007693 JAC51257.1 CP012528 ALC49635.1 CCAG010010280 GL763802 EFZ19272.1 GBXI01004143 JAD10149.1 CH916370 EDW00406.1 CH940651 EDW65330.1 AE014298 AL121804 AY119182 AAF45798.1 AAM51042.1 CAB65856.1 GALX01002315 JAB66151.1 ADTU01011299 CH933810 EDW07304.1 KQ976500 KYM83180.1 KQ979433 KYN21597.1 DS235073 EEB11392.1 GBHO01011172 GBHO01011171 JAG32432.1 JAG32433.1 GDHC01003081 JAQ15548.1 OUUW01000021 SPP89493.1 CH379063 EAL32628.2 GBHO01011177 GBHO01011170 GBRD01010021 JAG32427.1 JAG32434.1 JAG55803.1 SPP89492.1 GDHC01001000 JAQ17629.1 GDHC01006001 JAQ12628.1 CH963925 EDW78247.1 CH902632 EDV32840.1 KQ982373 KYQ57015.1 GBHO01011176 GBHO01011175 GBRD01010023 JAG32428.1 JAG32429.1 JAG55801.1 GL888932 EGI57185.1 CH480825 EDW44004.1 CH954183 EDV45642.1 KK856503 PTY25917.1 AK418526 BAN21653.1 LBMM01001677 KMQ95904.1 GFTR01003177 JAW13249.1 GFDF01007456 JAV06628.1 GFDF01007460 JAV06624.1 GEDC01018641 JAS18657.1 APGK01053845 BT127707 KB741237 KB629998 KB631549 AEE62669.1 ENN72189.1 ERL83326.1 ERL84371.1 GALX01002314 JAB66152.1 LNIX01000002 OXA60574.1 KA648518 AFP63147.1 GFDL01008488 JAV26557.1 DS231877 EDS42106.1 GFDL01008718 JAV26327.1 JRES01000819 KNC28153.1 ATLV01019037 KE525259 KFB43523.1 GGMS01012482 MBY81685.1 GL732524 EFX89776.1 AXCN02000238 AXCM01001682 AXCP01008404 AAAB01008879 EAA08411.4 APCN01002123
GDQN01000295 JAT85265.1 JAT88925.1 JAT90759.1 ODYU01007447 SOQ50228.1 GDQN01006837 JAT84217.1 NWSH01000010 PCG80931.1 GEBQ01025475 JAT14502.1 GECU01029102 JAS78604.1 KK852820 KDR15578.1 GECZ01004758 JAS65011.1 PYGN01000712 PSN41829.1 KZ288192 PBC34369.1 NEVH01007815 PNF35614.1 AAZX01000596 NNAY01004846 OXU17346.1 GBBI01000330 JAC18382.1 GEZM01054748 JAV73424.1 GEZM01054749 JAV73423.1 KQ977059 KYN06018.1 GL452364 EFN77405.1 LJIG01009419 KRT83117.1 LJIJ01000018 ODN05679.1 KK107459 QOIP01000004 EZA50387.1 RLU23713.1 KQ434827 KZC07497.1 GDKW01003552 JAI53043.1 GDHF01033525 JAI18789.1 KQ435794 KOX73640.1 GDHF01010467 GDHF01000405 JAI41847.1 JAI51909.1 UFQS01000668 UFQT01000668 SSX05971.1 SSX26328.1 KQ981673 KYN38370.1 GAMC01002542 JAC04014.1 GAKP01007699 GAKP01007697 GAKP01007695 GAKP01007693 JAC51257.1 CP012528 ALC49635.1 CCAG010010280 GL763802 EFZ19272.1 GBXI01004143 JAD10149.1 CH916370 EDW00406.1 CH940651 EDW65330.1 AE014298 AL121804 AY119182 AAF45798.1 AAM51042.1 CAB65856.1 GALX01002315 JAB66151.1 ADTU01011299 CH933810 EDW07304.1 KQ976500 KYM83180.1 KQ979433 KYN21597.1 DS235073 EEB11392.1 GBHO01011172 GBHO01011171 JAG32432.1 JAG32433.1 GDHC01003081 JAQ15548.1 OUUW01000021 SPP89493.1 CH379063 EAL32628.2 GBHO01011177 GBHO01011170 GBRD01010021 JAG32427.1 JAG32434.1 JAG55803.1 SPP89492.1 GDHC01001000 JAQ17629.1 GDHC01006001 JAQ12628.1 CH963925 EDW78247.1 CH902632 EDV32840.1 KQ982373 KYQ57015.1 GBHO01011176 GBHO01011175 GBRD01010023 JAG32428.1 JAG32429.1 JAG55801.1 GL888932 EGI57185.1 CH480825 EDW44004.1 CH954183 EDV45642.1 KK856503 PTY25917.1 AK418526 BAN21653.1 LBMM01001677 KMQ95904.1 GFTR01003177 JAW13249.1 GFDF01007456 JAV06628.1 GFDF01007460 JAV06624.1 GEDC01018641 JAS18657.1 APGK01053845 BT127707 KB741237 KB629998 KB631549 AEE62669.1 ENN72189.1 ERL83326.1 ERL84371.1 GALX01002314 JAB66152.1 LNIX01000002 OXA60574.1 KA648518 AFP63147.1 GFDL01008488 JAV26557.1 DS231877 EDS42106.1 GFDL01008718 JAV26327.1 JRES01000819 KNC28153.1 ATLV01019037 KE525259 KFB43523.1 GGMS01012482 MBY81685.1 GL732524 EFX89776.1 AXCN02000238 AXCM01001682 AXCP01008404 AAAB01008879 EAA08411.4 APCN01002123
Proteomes
UP000053268
UP000218220
UP000027135
UP000245037
UP000005203
UP000242457
+ More
UP000235965 UP000192223 UP000079169 UP000002358 UP000215335 UP000078542 UP000008237 UP000094527 UP000053097 UP000279307 UP000076502 UP000091820 UP000053105 UP000078541 UP000092553 UP000092445 UP000092444 UP000078200 UP000001070 UP000095301 UP000008792 UP000095300 UP000000803 UP000005205 UP000009192 UP000078540 UP000078492 UP000009046 UP000268350 UP000001819 UP000007798 UP000192221 UP000007801 UP000075809 UP000007755 UP000001292 UP000008711 UP000036403 UP000019118 UP000030742 UP000198287 UP000002320 UP000037069 UP000076408 UP000030765 UP000075881 UP000000305 UP000075886 UP000075883 UP000075880 UP000075885 UP000075901 UP000076407 UP000007062 UP000075840 UP000075902
UP000235965 UP000192223 UP000079169 UP000002358 UP000215335 UP000078542 UP000008237 UP000094527 UP000053097 UP000279307 UP000076502 UP000091820 UP000053105 UP000078541 UP000092553 UP000092445 UP000092444 UP000078200 UP000001070 UP000095301 UP000008792 UP000095300 UP000000803 UP000005205 UP000009192 UP000078540 UP000078492 UP000009046 UP000268350 UP000001819 UP000007798 UP000192221 UP000007801 UP000075809 UP000007755 UP000001292 UP000008711 UP000036403 UP000019118 UP000030742 UP000198287 UP000002320 UP000037069 UP000076408 UP000030765 UP000075881 UP000000305 UP000075886 UP000075883 UP000075880 UP000075885 UP000075901 UP000076407 UP000007062 UP000075840 UP000075902
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2W1BH21
A0A0N1IAH1
A0A1E1WPI6
A0A2H1WAY8
A0A1E1WBA5
A0A2A4K9M2
+ More
A0A1B6KSS0 A0A1B6HVA4 A0A067R0Z7 A0A1B6GRF3 A0A2P8YC43 A0A088AE92 A0A2A3ET98 A0A2J7R455 A0A1W4XS61 A0A1S3D724 K7JF93 A0A232EG66 A0A023FA80 A0A1Y1LIE7 A0A1Y1LIG9 A0A151IMA1 E2C419 A0A0T6B7S2 A0A1D2NKC8 A0A026W2T2 A0A154P6K5 A0A0P4VPG3 A0A0K8TWF7 A0A1A9X425 A0A0M8ZYE8 A0A0K8WLI6 A0A336MJT5 A0A195FCK4 W8C413 A0A034WAD4 A0A0M4EW59 A0A1A9ZFY6 A0A1B0FBL8 E9IJH8 A0A1A9V2Z0 A0A0A1XFG9 B4JLF3 A0A1I8N2L7 B4M3F5 A0A1I8PSZ1 Q9V3X4 V5I9U7 A0A158NBT0 B4L426 A0A195BG91 A0A151J9A0 E0VDD6 A0A0A9YMV8 A0A146M715 A0A3B0K831 Q29IM5 A0A0A9YH79 A0A3B0K9V8 A0A146MDX3 A0A146LYD4 B4N1K8 A0A1W4VTQ9 B3MYZ6 A0A151X9F0 A0A0A9YMV3 F4X8B5 B4I9X3 B3P937 A0A2R7X289 R4WEE9 A0A0J7L074 A0A224XYT7 A0A1L8DJI7 A0A1L8DJV7 A0A1B6CYY4 J3JWT9 V5GWQ7 A0A226EU41 T1PKQ2 A0A1Q3FG76 B0WB41 A0A1Q3FFK4 A0A0L0C6W5 A0A182XVJ4 A0A084VZX9 A0A2S2QVC6 A0A182K872 E9FSR3 A0A182QB60 A0A182MWD0 A0A182J0P3 A0A182PTM7 A0A182S6R6 A0A1Y9J159 Q7QBJ6 A0A182HJ35 A0A182UKU8
A0A1B6KSS0 A0A1B6HVA4 A0A067R0Z7 A0A1B6GRF3 A0A2P8YC43 A0A088AE92 A0A2A3ET98 A0A2J7R455 A0A1W4XS61 A0A1S3D724 K7JF93 A0A232EG66 A0A023FA80 A0A1Y1LIE7 A0A1Y1LIG9 A0A151IMA1 E2C419 A0A0T6B7S2 A0A1D2NKC8 A0A026W2T2 A0A154P6K5 A0A0P4VPG3 A0A0K8TWF7 A0A1A9X425 A0A0M8ZYE8 A0A0K8WLI6 A0A336MJT5 A0A195FCK4 W8C413 A0A034WAD4 A0A0M4EW59 A0A1A9ZFY6 A0A1B0FBL8 E9IJH8 A0A1A9V2Z0 A0A0A1XFG9 B4JLF3 A0A1I8N2L7 B4M3F5 A0A1I8PSZ1 Q9V3X4 V5I9U7 A0A158NBT0 B4L426 A0A195BG91 A0A151J9A0 E0VDD6 A0A0A9YMV8 A0A146M715 A0A3B0K831 Q29IM5 A0A0A9YH79 A0A3B0K9V8 A0A146MDX3 A0A146LYD4 B4N1K8 A0A1W4VTQ9 B3MYZ6 A0A151X9F0 A0A0A9YMV3 F4X8B5 B4I9X3 B3P937 A0A2R7X289 R4WEE9 A0A0J7L074 A0A224XYT7 A0A1L8DJI7 A0A1L8DJV7 A0A1B6CYY4 J3JWT9 V5GWQ7 A0A226EU41 T1PKQ2 A0A1Q3FG76 B0WB41 A0A1Q3FFK4 A0A0L0C6W5 A0A182XVJ4 A0A084VZX9 A0A2S2QVC6 A0A182K872 E9FSR3 A0A182QB60 A0A182MWD0 A0A182J0P3 A0A182PTM7 A0A182S6R6 A0A1Y9J159 Q7QBJ6 A0A182HJ35 A0A182UKU8
PDB
6MLU
E-value=3.25607e-56,
Score=550
Ontologies
GO
PANTHER
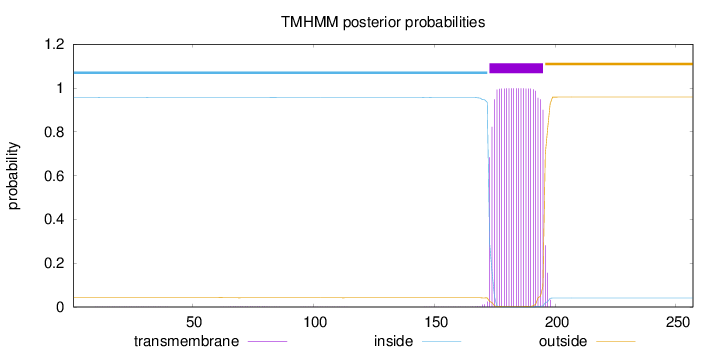
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
257
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.79686
Exp number, first 60 AAs:
0.01088
Total prob of N-in:
0.95649
inside
1 - 172
TMhelix
173 - 195
outside
196 - 257
Population Genetic Test Statistics
Pi
214.511374
Theta
159.868496
Tajima's D
1.503737
CLR
0.173677
CSRT
0.790710464476776
Interpretation
Uncertain
