Gene
KWMTBOMO14441
Pre Gene Modal
BGIBMGA000089
Annotation
PREDICTED:_uncharacterized_protein_LOC101746134_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.333
Sequence
CDS
ATGCGTAAAAAATCATTTCTTCTCGTCGTCTTCATTCAATTTTATTTTGCACAAACACATTATGGTAATGGTTTCTTCAAGGAGCAGAAGTCCTCTTACCGACCATACCAGCCAGTCTCCGTAAAGCATCACAAGGCTTACCCTCGACCTGAGGACAGATCAGACTATGATATCACGTCATACGACACGGAGTACGGCGCGGTCGTGTCAAACTTCACGTCTTCCATTTATGATGATAATCCTCAACTGTATCCTAATCCAGAGTTCATTGCGAACAGCAATCAAGGCAGTACTAAATTAAATGACAATAAAATATCCGACATAGAATTGCCAGATGACTCGTTAGCGGACGCAGACCTCAAAGAGCACGACATAATTGATAGCGTAACCTATCTATACGGATACAACAGTTGCCATGGCGATCGGCACGAATGGTTGATACTTATCTACGTAACAAGTGCTATAGCAGTAATAATACTGGCCGGCGCGGTAATGTGGCAGATGTGGAGCAATTACGCGGCGGAATTCAGAATGAAGAGTAAACTGTATCCTATCAACATCAATCTGTGCTGTTGTCTAGGAGTCTGCACTCTCATTTACATACAGGCTGTTTTGGGCGTGTCCTCGCCTTCTCAATGCGAGAGGATCGCCCTGCTTCTCCACTACACGCATATAACTTGCGCGATGTGGATCGTAGCCTTGGCCGCGGCCGTCGCAGAATATTGCACGTGCGATACCCTATTGCCTCTCAAGTACAACTACCTACTGGCCTACGGCGTTCCTGCCGTAGTTGTTATGTTCAACTACGCCCTGTCAATGGAACATTATGAAATAAAGCATTACTGCTGGATGTCTATCGAGAAAGGCATGGTCATGGGCTTTATGGTACCAGCGATGATTCTCATTTTAATTAATACTGGCATCGTGATACTAGGCCTAAAGAGCGTGAATAAAAAACAAGCGGAAATGCTGACGGCCAAGATACAAGAACTCGTGGACCACCACATTACGAACTGGCCAAAGAATGATCAAAACGGCGATAAAAATGCAAGTATTGAAACCTTGGATAATATTTACACGCCGAGCAGCAGTAGAAAGAATACTGACAGTTCGGAAACTCTCGATAAAGACTACAATGCCAGCGAAGAAGATTATAATTATTCTAATAACGGAGTAGCGAACGATGAAGCTGGTGAAAATCCGATTGAGCGATCCGCTTCCAATAAAACAATACAGGACAAAAGCCTGTTAAATATGCTGTATATGGATAACATTTCGTTAAAATGGAGTTGGAACACGGAAGGCAATGAATTGAAGACATATTTGAATTTATGTTTGATTTTGGAGCCATTTTTCGCCATCAACTGGGTTATGGGCGTTGTTGCTATAGAGAATGCCACTCACTGGTCGACCCCGACGATTTATTTAATACTAATTGTGTCTATGTACATGTACTTGACTGGTACTATATGTACCACCTTGCCTATAGTACAACAGAAGACTCCCGTGCCGCCGTGCTGTGAAGACGTTGTGATTGAACCTTTCCTAACAAGAACAAGGACAACTGACAGCATTCCACTTTTGGACCCGACTGTACAGCAGCCAAACGTAACTCCAGCACCTGCGGACACTATTAGCACAATTAGTATTTAG
Protein
MRKKSFLLVVFIQFYFAQTHYGNGFFKEQKSSYRPYQPVSVKHHKAYPRPEDRSDYDITSYDTEYGAVVSNFTSSIYDDNPQLYPNPEFIANSNQGSTKLNDNKISDIELPDDSLADADLKEHDIIDSVTYLYGYNSCHGDRHEWLILIYVTSAIAVIILAGAVMWQMWSNYAAEFRMKSKLYPININLCCCLGVCTLIYIQAVLGVSSPSQCERIALLLHYTHITCAMWIVALAAAVAEYCTCDTLLPLKYNYLLAYGVPAVVVMFNYALSMEHYEIKHYCWMSIEKGMVMGFMVPAMILILINTGIVILGLKSVNKKQAEMLTAKIQELVDHHITNWPKNDQNGDKNASIETLDNIYTPSSSRKNTDSSETLDKDYNASEEDYNYSNNGVANDEAGENPIERSASNKTIQDKSLLNMLYMDNISLKWSWNTEGNELKTYLNLCLILEPFFAINWVMGVVAIENATHWSTPTIYLILIVSMYMYLTGTICTTLPIVQQKTPVPPCCEDVVIEPFLTRTRTTDSIPLLDPTVQQPNVTPAPADTISTISI
Summary
Similarity
Belongs to the FGGY kinase family.
Uniprot
Pubmed
EMBL
BABH01024190
KZ150182
PZC72499.1
NWSH01000010
PCG80934.1
ODYU01008109
+ More
SOQ51424.1 GDQN01008837 GDQN01008583 GDQN01001044 JAT82217.1 JAT82471.1 JAT90010.1 AGBW02014229 OWR42048.1 KQ458995 KPJ04406.1 KQ460195 KPJ17213.1 JTDY01007158 KOB65425.1 ADMH02001287 ETN63154.1 AXCN02001151 DS234266 EDS33435.1 JXUM01052188 KQ561724 KXJ77715.1 AJ439353 AAAB01008987 CAD27928.1 EAA01366.2 APCN01000883 AXCM01006311 ATLV01003738 ATLV01003739 KE524159 KFB34969.1
SOQ51424.1 GDQN01008837 GDQN01008583 GDQN01001044 JAT82217.1 JAT82471.1 JAT90010.1 AGBW02014229 OWR42048.1 KQ458995 KPJ04406.1 KQ460195 KPJ17213.1 JTDY01007158 KOB65425.1 ADMH02001287 ETN63154.1 AXCN02001151 DS234266 EDS33435.1 JXUM01052188 KQ561724 KXJ77715.1 AJ439353 AAAB01008987 CAD27928.1 EAA01366.2 APCN01000883 AXCM01006311 ATLV01003738 ATLV01003739 KE524159 KFB34969.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000000673 UP000069272 UP000075880 UP000075881 UP000075901 UP000075886 UP000075900 UP000002320 UP000075902 UP000075920 UP000069940 UP000249989 UP000007062 UP000075885 UP000075840 UP000075903 UP000075884 UP000075883 UP000030765
UP000000673 UP000069272 UP000075880 UP000075881 UP000075901 UP000075886 UP000075900 UP000002320 UP000075902 UP000075920 UP000069940 UP000249989 UP000007062 UP000075885 UP000075840 UP000075903 UP000075884 UP000075883 UP000030765
PRIDE
Interpro
ProteinModelPortal
Ontologies
GO
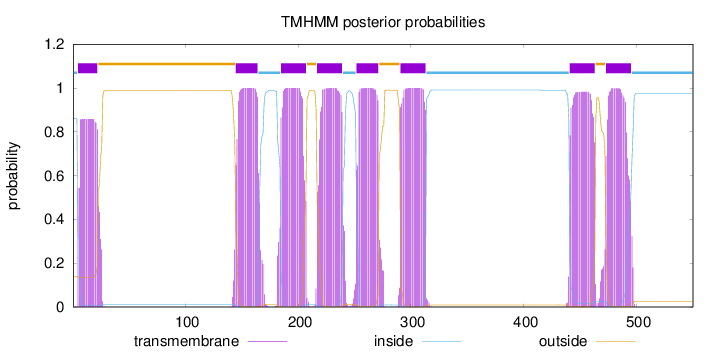
Topology
SignalP
Position: 1 - 24,
Likelihood: 0.747021

Length:
550
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
169.83402
Exp number, first 60 AAs:
16.23359
Total prob of N-in:
0.86064
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 22
outside
23 - 144
TMhelix
145 - 164
inside
165 - 184
TMhelix
185 - 207
outside
208 - 216
TMhelix
217 - 239
inside
240 - 251
TMhelix
252 - 271
outside
272 - 290
TMhelix
291 - 313
inside
314 - 440
TMhelix
441 - 463
outside
464 - 472
TMhelix
473 - 495
inside
496 - 550
Population Genetic Test Statistics
Pi
273.484212
Theta
165.362265
Tajima's D
1.40419
CLR
0.316382
CSRT
0.767961601919904
Interpretation
Uncertain
