Gene
KWMTBOMO14434
Pre Gene Modal
BGIBMGA000093
Annotation
sex_peptide_receptor_[Bombyx_mori]
Full name
Sex peptide receptor
Location in the cell
PlasmaMembrane Reliability : 4.89
Sequence
CDS
ATGGCGGTCACCATAGACAATTCAACGAACGACTTCGAATTCCAGAAGCCATTCAACTACTCAATTAACGAGAACATCACCTACTTCGATTACACGAACTTCACTTCGGACGATTTCTGCGCATCAAACAACTCACACGTATACTTAAACGTCACCTGTGAGTTCGCTATAAGCTATGCTGAGCCTATGTACGGGTACATCGCACCGTTTCTGCTAGCTACAACGACAGTTGCCAACACCCTGATCGTGGTGGTGCTATCGCGGAGGCACATGAGGACGCCCACCAATGCCGTGCTGATGGCGATGGCTCTTTGCGATATGTTCACCATGCTGTTCCCGGCGCCGTGGCTATTCTACATGTACACTTTTGGAAACCACTACAAGCCGCTAAGTCCCGTCAGAGCCTGCCAAGCTTGGAATTACATGAACGAGGTCATTCCAGCGATGTTCCACACGGCTAGTATCTGGTTGACGTTAGCCTTAGCCGTCCAGAGATATATCTACGTTTGCCACGCGCCCGTCGCCAGGACCTGGTGTACCATGCCCCGGGTGATGAAATGTCTGATCTACATAGGAATAGCGGCGTTTCTTCATCAGCTACCGCGGTTCTTCGACAGATGCTACACTCCCCACAAGACTGTGTGGCGGGGGCGCGTGGAGGAAGTCTGCAGAATCGAGATGGCGTCTTGGGTGAAAGCGCTCTCCGTCGACGCCTATTTCATCTCCTACTTCGGCTTCAGAGTTTTGTTCGTCCACTTAATCCCGTGCACATCTTTAGTTGTCCTGAACGTGCTCCTCTTCAGGGCCATGAGGACGGCCCAAATAAACAGACAGAAGCTGTTCAAAGAAAACCGAAAATCGGAGTGCAAGAGACTCAGAGATTCTAACTGCACCACCCTGATGTTGATAGTCGTGGTCACGGTTTTCCTCCTCGTGGAAATACCCGTGGCCGTCGTCACTATCCTCCACATAATATCCAGCACGATTGTCGAGATCTTGGACTACCATATCGCGAATATTCTCGTTCTGGTAACGAATTTCTTCATTATCGTTTCCTATCCAATCAACTTTGCTATATATTGCGGTATGTCGCGTCAATTCAGGGAGACCTTCAAAGAATTGTTTATCAGAGGCACGGTTACTAGTAGGAAGAACGGTGGCTCCAGTAGGTACTCTCTTGTCAATGGCCCGAGGACCTGTACAAACGAAACTGTGCTTTAA
Protein
MAVTIDNSTNDFEFQKPFNYSINENITYFDYTNFTSDDFCASNNSHVYLNVTCEFAISYAEPMYGYIAPFLLATTTVANTLIVVVLSRRHMRTPTNAVLMAMALCDMFTMLFPAPWLFYMYTFGNHYKPLSPVRACQAWNYMNEVIPAMFHTASIWLTLALAVQRYIYVCHAPVARTWCTMPRVMKCLIYIGIAAFLHQLPRFFDRCYTPHKTVWRGRVEEVCRIEMASWVKALSVDAYFISYFGFRVLFVHLIPCTSLVVLNVLLFRAMRTAQINRQKLFKENRKSECKRLRDSNCTTLMLIVVVTVFLLVEIPVAVVTILHIISSTIVEILDYHIANILVLVTNFFIIVSYPINFAIYCGMSRQFRETFKELFIRGTVTSRKNGGSSRYSLVNGPRTCTNETVL
Summary
Description
Receptor for two functionally unrelated ligands; SP (A70A) for controlling reproductive behaviors and MIP for controlling sleep behavior (PubMed:18066048, PubMed:20458515, PubMed:20308537, PubMed:24089336, PubMed:25333796). MIP-SPR pathway functions as a sleep homeostat which perceives the need for sleep and stabilizes it by providing a slow-acting inhibitory input to the fly arousal system that involve the pigment dispersing factor (pdf) neurons (PubMed:25333796). SP-SPR is one of the multiple SP pathways that induce female post-mating behavioral responses (PMR) such as the suppression of mating receptivity and initiation of egg laying (PubMed:18066048, PubMed:24089336). The PMR switch is achieved by mediating the synaptic output of neurons such as those expressing fruitless (fru), double sex (dsx) and pickpocket (ppk) (PubMed:18066048, PubMed:24089336).
Similarity
Belongs to the G-protein coupled receptor 1 family.
Keywords
Cell membrane
Complete proteome
G-protein coupled receptor
Membrane
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain Sex peptide receptor
Uniprot
B0F4F1
M1KBS6
A0A0S1YDF7
I0AW25
E0A919
A0A0N1I6P7
+ More
A0A194QE30 A0A212FD50 A0A1S3CY93 A0A1B6JXI3 A0A1B6LE68 G0ZGH2 A0A2J7RB67 A0A067RJM6 A0A023F5Z2 A0A2S2QQC0 A0A2P8ZJS8 U3U7M1 A0A2H8TQQ4 J9JSJ4 T1H9A3 G0ZGH1 A0A1Y1LIF7 E0VDC2 B0F4F2 A0A1B6DZV1 A0A1L8DK97 B0F4E9 Q29I83 A0A3B0J9B4 A0A182RD27 B4PZP9 A0A182FB29 B3MR14 B4NCZ3 B4JN65 B4M2J2 B4I0P1 B3NV47 B4R4Y8 Q8SWR3 B4L4M1 A0A1W4WUI1 A0A182JY91 A0A1W4VSA3 B0F4F0 A0A0M4EMH0 A0A1Y9IVJ8 A0A1L8DK23 A0A0L0C1L1 A0A1I8PMA8 A0A1B0ADX5 A0A1A9VLH6 A0A226EVS0 A0A1B0ANI1 A0A1I8MLC0 A0A1D2NN16 A0A034VH36 A0A0K8U8G9 A0A0T6BDH4 A0A1A9WL69 A0A087SZ34 W8C929 A0A0A1WWG3 A0A2L2YMB8 A0A0P4Z222 A0A0P5JPP2 A0A0P5TP65 A0A162CMJ5 W5JIG6 E9GCL6 A0A131Z615 A0A224YT96 L7MG47 L7MG28 A0A131XKL4 A0A1E1XKG9 A0A1E1X6I3 L7ZCA0 A0A0K8RRP0 L7ZA74 A0A0P6I0U9 A0A3R7M1D8 A0A2P6L8W4 A0A0P6E469 A0A1Z5L7P5 A0A293MAE0 B7PXI7 A0A293LM40 A0A1B0GIR2 A0A0P6CQU4 L7MGG5 A0A1J1HJE3 A0A182GS94 A0A0P5TKZ5 A0A182LTG4
A0A194QE30 A0A212FD50 A0A1S3CY93 A0A1B6JXI3 A0A1B6LE68 G0ZGH2 A0A2J7RB67 A0A067RJM6 A0A023F5Z2 A0A2S2QQC0 A0A2P8ZJS8 U3U7M1 A0A2H8TQQ4 J9JSJ4 T1H9A3 G0ZGH1 A0A1Y1LIF7 E0VDC2 B0F4F2 A0A1B6DZV1 A0A1L8DK97 B0F4E9 Q29I83 A0A3B0J9B4 A0A182RD27 B4PZP9 A0A182FB29 B3MR14 B4NCZ3 B4JN65 B4M2J2 B4I0P1 B3NV47 B4R4Y8 Q8SWR3 B4L4M1 A0A1W4WUI1 A0A182JY91 A0A1W4VSA3 B0F4F0 A0A0M4EMH0 A0A1Y9IVJ8 A0A1L8DK23 A0A0L0C1L1 A0A1I8PMA8 A0A1B0ADX5 A0A1A9VLH6 A0A226EVS0 A0A1B0ANI1 A0A1I8MLC0 A0A1D2NN16 A0A034VH36 A0A0K8U8G9 A0A0T6BDH4 A0A1A9WL69 A0A087SZ34 W8C929 A0A0A1WWG3 A0A2L2YMB8 A0A0P4Z222 A0A0P5JPP2 A0A0P5TP65 A0A162CMJ5 W5JIG6 E9GCL6 A0A131Z615 A0A224YT96 L7MG47 L7MG28 A0A131XKL4 A0A1E1XKG9 A0A1E1X6I3 L7ZCA0 A0A0K8RRP0 L7ZA74 A0A0P6I0U9 A0A3R7M1D8 A0A2P6L8W4 A0A0P6E469 A0A1Z5L7P5 A0A293MAE0 B7PXI7 A0A293LM40 A0A1B0GIR2 A0A0P6CQU4 L7MGG5 A0A1J1HJE3 A0A182GS94 A0A0P5TKZ5 A0A182LTG4
Pubmed
18066048
21426940
26354079
22118469
24845553
25474469
+ More
29403074 23932938 25218475 25401762 28004739 20566863 18362917 19820115 15632085 17994087 23185243 17550304 18057021 10731132 12537572 12537569 20458515 20308537 24089336 25333796 12364791 26108605 25315136 27289101 25348373 24495485 25830018 26561354 20920257 23761445 21292972 26830274 28797301 25576852 28049606 29209593 28503490 23357681 28528879 26483478
29403074 23932938 25218475 25401762 28004739 20566863 18362917 19820115 15632085 17994087 23185243 17550304 18057021 10731132 12537572 12537569 20458515 20308537 24089336 25333796 12364791 26108605 25315136 27289101 25348373 24495485 25830018 26561354 20920257 23761445 21292972 26830274 28797301 25576852 28049606 29209593 28503490 23357681 28528879 26483478
EMBL
EU106876
ABW86946.1
JX070670
AGE92037.1
KT031042
ALM88340.1
+ More
JQ689079 AFH53182.1 HM567403 ADK79103.2 KQ461174 KPJ08003.1 KQ459144 KPJ03732.1 AGBW02009107 OWR51664.1 MG550199 AWT50633.1 GECU01007835 GECU01003817 JAS99871.1 JAT03890.1 GEBQ01017992 JAT21985.1 JF273643 AEK80440.1 NEVH01006563 PNF38070.1 KK852431 KDR24017.1 GBBI01002107 JAC16605.1 GGMS01010527 MBY79730.1 PYGN01000035 PSN56754.1 AB817293 BAO01060.1 GFXV01004722 MBW16527.1 ABLF02040252 ABLF02040268 ABLF02040269 ABLF02040270 ACPB03007534 KF958188 AIQ81191.1 JF273642 GBHO01025389 AEK80439.1 JAG18215.1 GEZM01054743 GEZM01054742 JAV73429.1 DS235073 EEB11378.1 EU106877 EU443244 KQ971321 ABW86947.1 ACC68841.1 EFA01285.2 GEDC01024257 GEDC01016961 GEDC01006108 JAS13041.1 JAS20337.1 JAS31190.1 GFDF01007287 JAV06797.1 EU106874 ABW86944.1 EU106873 CH379063 ABW86943.1 EAL32770.3 KRT06310.1 KRT06311.1 OUUW01000003 SPP78854.1 CM000162 EDX01116.1 KRK05960.1 KRK05961.1 KRK05962.1 KRK05963.1 CH902622 EDV34219.1 KPU75025.1 CH964239 EDW82702.2 CH916371 EDV92158.1 CH940651 EDW65896.1 CH480819 EDW53072.1 CH954180 EDV45895.1 CM000366 EDX17137.1 EU443243 AE014298 AY095526 AAM12257.1 ACC68840.1 CH933810 EDW07499.2 AAAB01008986 EU106875 ABW86945.1 CP012528 ALC48934.1 GFDF01007298 JAV06786.1 JRES01000997 KNC26253.1 LNIX01000002 OXA60696.1 JXJN01000834 LJIJ01000001 ODN06668.1 GAKP01016336 JAC42616.1 GDHF01029360 GDHF01017219 JAI22954.1 JAI35095.1 LJIG01001646 KRT85294.1 KK112619 KFM58123.1 GAMC01002519 JAC04037.1 GBXI01011462 GBXI01005475 JAD02830.1 JAD08817.1 IAAA01036746 IAAA01036747 LAA09243.1 GDIP01231237 JAI92164.1 GDIQ01201716 GDIP01114495 JAK50009.1 JAL89219.1 GDIP01126836 JAL76878.1 LRGB01000944 KZS14576.1 ADMH02001383 ETN62690.1 GL732539 EFX82580.1 GEDV01002332 JAP86225.1 GFPF01007823 MAA18969.1 GACK01002796 JAA62238.1 GACK01002247 JAA62787.1 GEFH01002375 JAP66206.1 GFAA01003614 JAT99820.1 GFAC01004309 JAT94879.1 KC422390 AGE11604.1 GADI01000514 JAA73294.1 KC422391 AGE11605.1 GDIQ01011073 JAN83664.1 QCYY01002449 ROT70219.1 MWRG01000907 PRD35009.1 GDIQ01069216 JAN25521.1 GFJQ02003523 JAW03447.1 GFWV01017979 MAA42707.1 ABJB010212851 ABJB010934302 DS814451 EEC11309.1 GFWV01004181 MAA28911.1 AJWK01015325 AJWK01015326 AJWK01015327 AJWK01015328 AJWK01015329 AJWK01015330 AJWK01015331 GDIP01010477 JAM93238.1 GACK01002671 JAA62363.1 CVRI01000006 CRK88141.1 JXUM01016857 JXUM01016858 JXUM01016859 JXUM01016860 JXUM01016861 KQ560467 KXJ82271.1 GDIP01128326 JAL75388.1 AXCM01012874
JQ689079 AFH53182.1 HM567403 ADK79103.2 KQ461174 KPJ08003.1 KQ459144 KPJ03732.1 AGBW02009107 OWR51664.1 MG550199 AWT50633.1 GECU01007835 GECU01003817 JAS99871.1 JAT03890.1 GEBQ01017992 JAT21985.1 JF273643 AEK80440.1 NEVH01006563 PNF38070.1 KK852431 KDR24017.1 GBBI01002107 JAC16605.1 GGMS01010527 MBY79730.1 PYGN01000035 PSN56754.1 AB817293 BAO01060.1 GFXV01004722 MBW16527.1 ABLF02040252 ABLF02040268 ABLF02040269 ABLF02040270 ACPB03007534 KF958188 AIQ81191.1 JF273642 GBHO01025389 AEK80439.1 JAG18215.1 GEZM01054743 GEZM01054742 JAV73429.1 DS235073 EEB11378.1 EU106877 EU443244 KQ971321 ABW86947.1 ACC68841.1 EFA01285.2 GEDC01024257 GEDC01016961 GEDC01006108 JAS13041.1 JAS20337.1 JAS31190.1 GFDF01007287 JAV06797.1 EU106874 ABW86944.1 EU106873 CH379063 ABW86943.1 EAL32770.3 KRT06310.1 KRT06311.1 OUUW01000003 SPP78854.1 CM000162 EDX01116.1 KRK05960.1 KRK05961.1 KRK05962.1 KRK05963.1 CH902622 EDV34219.1 KPU75025.1 CH964239 EDW82702.2 CH916371 EDV92158.1 CH940651 EDW65896.1 CH480819 EDW53072.1 CH954180 EDV45895.1 CM000366 EDX17137.1 EU443243 AE014298 AY095526 AAM12257.1 ACC68840.1 CH933810 EDW07499.2 AAAB01008986 EU106875 ABW86945.1 CP012528 ALC48934.1 GFDF01007298 JAV06786.1 JRES01000997 KNC26253.1 LNIX01000002 OXA60696.1 JXJN01000834 LJIJ01000001 ODN06668.1 GAKP01016336 JAC42616.1 GDHF01029360 GDHF01017219 JAI22954.1 JAI35095.1 LJIG01001646 KRT85294.1 KK112619 KFM58123.1 GAMC01002519 JAC04037.1 GBXI01011462 GBXI01005475 JAD02830.1 JAD08817.1 IAAA01036746 IAAA01036747 LAA09243.1 GDIP01231237 JAI92164.1 GDIQ01201716 GDIP01114495 JAK50009.1 JAL89219.1 GDIP01126836 JAL76878.1 LRGB01000944 KZS14576.1 ADMH02001383 ETN62690.1 GL732539 EFX82580.1 GEDV01002332 JAP86225.1 GFPF01007823 MAA18969.1 GACK01002796 JAA62238.1 GACK01002247 JAA62787.1 GEFH01002375 JAP66206.1 GFAA01003614 JAT99820.1 GFAC01004309 JAT94879.1 KC422390 AGE11604.1 GADI01000514 JAA73294.1 KC422391 AGE11605.1 GDIQ01011073 JAN83664.1 QCYY01002449 ROT70219.1 MWRG01000907 PRD35009.1 GDIQ01069216 JAN25521.1 GFJQ02003523 JAW03447.1 GFWV01017979 MAA42707.1 ABJB010212851 ABJB010934302 DS814451 EEC11309.1 GFWV01004181 MAA28911.1 AJWK01015325 AJWK01015326 AJWK01015327 AJWK01015328 AJWK01015329 AJWK01015330 AJWK01015331 GDIP01010477 JAM93238.1 GACK01002671 JAA62363.1 CVRI01000006 CRK88141.1 JXUM01016857 JXUM01016858 JXUM01016859 JXUM01016860 JXUM01016861 KQ560467 KXJ82271.1 GDIP01128326 JAL75388.1 AXCM01012874
Proteomes
UP000053240
UP000053268
UP000007151
UP000079169
UP000235965
UP000027135
+ More
UP000245037 UP000007819 UP000015103 UP000009046 UP000007266 UP000001819 UP000268350 UP000075900 UP000002282 UP000069272 UP000007801 UP000007798 UP000001070 UP000008792 UP000001292 UP000008711 UP000000304 UP000000803 UP000009192 UP000192223 UP000075881 UP000192221 UP000092553 UP000075920 UP000037069 UP000095300 UP000092445 UP000078200 UP000198287 UP000092460 UP000095301 UP000094527 UP000091820 UP000054359 UP000076858 UP000000673 UP000000305 UP000283509 UP000001555 UP000092461 UP000183832 UP000069940 UP000249989 UP000075883
UP000245037 UP000007819 UP000015103 UP000009046 UP000007266 UP000001819 UP000268350 UP000075900 UP000002282 UP000069272 UP000007801 UP000007798 UP000001070 UP000008792 UP000001292 UP000008711 UP000000304 UP000000803 UP000009192 UP000192223 UP000075881 UP000192221 UP000092553 UP000075920 UP000037069 UP000095300 UP000092445 UP000078200 UP000198287 UP000092460 UP000095301 UP000094527 UP000091820 UP000054359 UP000076858 UP000000673 UP000000305 UP000283509 UP000001555 UP000092461 UP000183832 UP000069940 UP000249989 UP000075883
Interpro
ProteinModelPortal
B0F4F1
M1KBS6
A0A0S1YDF7
I0AW25
E0A919
A0A0N1I6P7
+ More
A0A194QE30 A0A212FD50 A0A1S3CY93 A0A1B6JXI3 A0A1B6LE68 G0ZGH2 A0A2J7RB67 A0A067RJM6 A0A023F5Z2 A0A2S2QQC0 A0A2P8ZJS8 U3U7M1 A0A2H8TQQ4 J9JSJ4 T1H9A3 G0ZGH1 A0A1Y1LIF7 E0VDC2 B0F4F2 A0A1B6DZV1 A0A1L8DK97 B0F4E9 Q29I83 A0A3B0J9B4 A0A182RD27 B4PZP9 A0A182FB29 B3MR14 B4NCZ3 B4JN65 B4M2J2 B4I0P1 B3NV47 B4R4Y8 Q8SWR3 B4L4M1 A0A1W4WUI1 A0A182JY91 A0A1W4VSA3 B0F4F0 A0A0M4EMH0 A0A1Y9IVJ8 A0A1L8DK23 A0A0L0C1L1 A0A1I8PMA8 A0A1B0ADX5 A0A1A9VLH6 A0A226EVS0 A0A1B0ANI1 A0A1I8MLC0 A0A1D2NN16 A0A034VH36 A0A0K8U8G9 A0A0T6BDH4 A0A1A9WL69 A0A087SZ34 W8C929 A0A0A1WWG3 A0A2L2YMB8 A0A0P4Z222 A0A0P5JPP2 A0A0P5TP65 A0A162CMJ5 W5JIG6 E9GCL6 A0A131Z615 A0A224YT96 L7MG47 L7MG28 A0A131XKL4 A0A1E1XKG9 A0A1E1X6I3 L7ZCA0 A0A0K8RRP0 L7ZA74 A0A0P6I0U9 A0A3R7M1D8 A0A2P6L8W4 A0A0P6E469 A0A1Z5L7P5 A0A293MAE0 B7PXI7 A0A293LM40 A0A1B0GIR2 A0A0P6CQU4 L7MGG5 A0A1J1HJE3 A0A182GS94 A0A0P5TKZ5 A0A182LTG4
A0A194QE30 A0A212FD50 A0A1S3CY93 A0A1B6JXI3 A0A1B6LE68 G0ZGH2 A0A2J7RB67 A0A067RJM6 A0A023F5Z2 A0A2S2QQC0 A0A2P8ZJS8 U3U7M1 A0A2H8TQQ4 J9JSJ4 T1H9A3 G0ZGH1 A0A1Y1LIF7 E0VDC2 B0F4F2 A0A1B6DZV1 A0A1L8DK97 B0F4E9 Q29I83 A0A3B0J9B4 A0A182RD27 B4PZP9 A0A182FB29 B3MR14 B4NCZ3 B4JN65 B4M2J2 B4I0P1 B3NV47 B4R4Y8 Q8SWR3 B4L4M1 A0A1W4WUI1 A0A182JY91 A0A1W4VSA3 B0F4F0 A0A0M4EMH0 A0A1Y9IVJ8 A0A1L8DK23 A0A0L0C1L1 A0A1I8PMA8 A0A1B0ADX5 A0A1A9VLH6 A0A226EVS0 A0A1B0ANI1 A0A1I8MLC0 A0A1D2NN16 A0A034VH36 A0A0K8U8G9 A0A0T6BDH4 A0A1A9WL69 A0A087SZ34 W8C929 A0A0A1WWG3 A0A2L2YMB8 A0A0P4Z222 A0A0P5JPP2 A0A0P5TP65 A0A162CMJ5 W5JIG6 E9GCL6 A0A131Z615 A0A224YT96 L7MG47 L7MG28 A0A131XKL4 A0A1E1XKG9 A0A1E1X6I3 L7ZCA0 A0A0K8RRP0 L7ZA74 A0A0P6I0U9 A0A3R7M1D8 A0A2P6L8W4 A0A0P6E469 A0A1Z5L7P5 A0A293MAE0 B7PXI7 A0A293LM40 A0A1B0GIR2 A0A0P6CQU4 L7MGG5 A0A1J1HJE3 A0A182GS94 A0A0P5TKZ5 A0A182LTG4
PDB
6ME8
E-value=4.93206e-07,
Score=129
Ontologies
GO
Topology
Subcellular location
Cell membrane
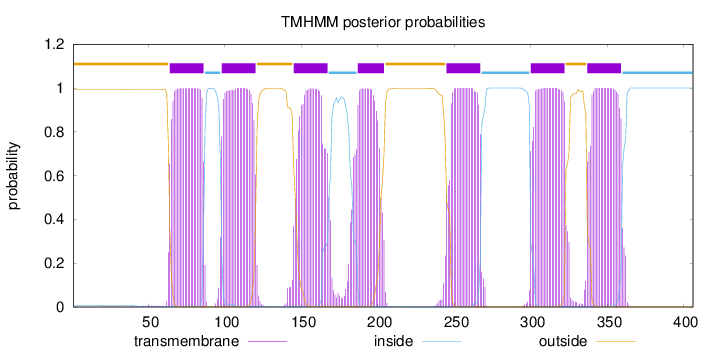
Length:
406
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
154.40645
Exp number, first 60 AAs:
0.09633
Total prob of N-in:
0.00756
outside
1 - 63
TMhelix
64 - 86
inside
87 - 97
TMhelix
98 - 120
outside
121 - 144
TMhelix
145 - 167
inside
168 - 186
TMhelix
187 - 204
outside
205 - 244
TMhelix
245 - 267
inside
268 - 299
TMhelix
300 - 322
outside
323 - 336
TMhelix
337 - 359
inside
360 - 406
Population Genetic Test Statistics
Pi
260.069708
Theta
180.621133
Tajima's D
1.620935
CLR
0.256601
CSRT
0.817159142042898
Interpretation
Uncertain
