Gene
KWMTBOMO14429
Pre Gene Modal
BGIBMGA000125
Annotation
PREDICTED:_uncharacterized_protein_LOC105390855_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.243
Sequence
CDS
ATGGTTCGAATTACAACTTACGTATTTTTCGACATCGAAACAACAGGTTTGCCGTTTCAAGAAAGAAATAAAACTAAGATCACCGAATTATGTTTTTTGGCAGTGTCTAGAGATGATCTAGATATACAATCAGGTCTGCCACCCATTAAGAAGTTGTCTTTCGTACTGAACCCAGAAAAGAAAATTCATCCTGATGTTGTAGTTTTGACTGGCCTAACGAATGAGTCACTAAAGAATGCGCCGACATTTAAACAGAGAGCAAATGCTATAGTATCTTTTTTAAATGAACTTCCGAAGCCTATATGTTTAGTCGCGCATAATGGAAATTCATTCGATTATAAGATATTATTAGCAGAGTGCAACGATGCAGGCATTTTGTTGCCAAGTGACTTACTGTGCATAGACTCTTTGATTGGCTTCCGAAAAATTCTAAAGACCACTCAGAAGTCATTACCATTAAAAATGAATACTAGCAAAGACGATAGTGCTAGTGAATTGCTATGGCCAGAACTCGACGTGTCTACAGAAAATTGGGAAGAAATTGATAATCTGTGTGCCTCATTTAGCAAAGACACATGTATTGATGATGAAAAAAGTAAAAAAGAAAATGATTTAAATCAGCAAAAGTGTACGAATTTAAAGAAGGGTCGGAAAAATAAGAATGACCTTACAACTCAAGATCAAAGTTTGTCAATACAAAATAAAAGTACTAAAAAGAGGGAGTTTACTTTGACTGGTTTATATAAAAGACTACTTGATAAAGATGCAGTAAATGCACACAGAGCAGAAGATGACTGTGTTATACTTATAGAATGTGTATTGGCTCTACGAGATGAGTTCTTGACATGGGCTGATGAGTCTTGTAAAACAATACACCAAATCAAGCCTTTAATACGCTATTAG
Protein
MVRITTYVFFDIETTGLPFQERNKTKITELCFLAVSRDDLDIQSGLPPIKKLSFVLNPEKKIHPDVVVLTGLTNESLKNAPTFKQRANAIVSFLNELPKPICLVAHNGNSFDYKILLAECNDAGILLPSDLLCIDSLIGFRKILKTTQKSLPLKMNTSKDDSASELLWPELDVSTENWEEIDNLCASFSKDTCIDDEKSKKENDLNQQKCTNLKKGRKNKNDLTTQDQSLSIQNKSTKKREFTLTGLYKRLLDKDAVNAHRAEDDCVILIECVLALRDEFLTWADESCKTIHQIKPLIRY
Summary
Uniprot
H9IS98
A0A2A4JRW8
A0A2H1WDQ2
A0A194Q5X9
A0A0L7KPL8
S4PLU4
+ More
A0A0N0PES1 A0A1Y1MQH1 S4PXS2 A0A182PMH4 D6WI30 A0A0N1IIR4 A0A182SXW8 A0A194Q5D3 A0A1B6FXI8 A0A0T6BBG6 A0A182RJN9 A0A0L7KQY3 A0A2A4K8M0 A0A139WK20 N6T731 A0A182Q4Y8 A0A182Y2E6 A0A182JHX3 A0A084WK02 A0A1B6L8W3 A0A182N976 J3JX86 A0A1S3CUY4 A0A1B6JLD2 E0W1R9 A0A067RLU5 A0A182VMN9 A0A182TV85 A0A182I4P2 Q5TQT2 A0A154PLK4 A0A0Q9XC88 B4KHL1 A0A182X4P5 A0A1I8N3X4 B7PN33 A0A182LK68 A0A182FDU9 A0A0L7R9E7 A0A1B6EAM9 A0A1W4V6M9 A0A2W1BL26 A0A0L8G521 A0A1Z5KVU7 H9JVW3 A0A232FJI8 A0A088A5Q7 V5HZ96 K7J083 B4N101 A0A023F6M6 A0A0K8W4C0 A0A0V0GB29 A0A1I8PB84 A0A1Q3FPV4 A0A034WIT6 B4M9B2 A0A1I8PQ47 A0A2A3E511 U5EQW5 B4JCS3 A0A151I4G8 A0A0J7KIH0 A0A131Z5C4 A0A151WQP9 A0A1S4EUT2 Q0C7B5 A0A195D0Q0 A0A0A1WSR5 A0A069DYI9 A0A182GUN5 B0WAN4 A0A023F8D4 A0A224XS15 A0A2P6KP25 A0A1I8MAA6 A0A2W1BSK7 Q9VQJ7 A0A0V0GCM6 B3NAN4 E1ZUW4 A0A0P5TK85 A0A2A4J5B2 A0A195FTI0 B4I2T4 B4NXB4 A0A0P5JYQ5 A0A0A9XWM0
A0A0N0PES1 A0A1Y1MQH1 S4PXS2 A0A182PMH4 D6WI30 A0A0N1IIR4 A0A182SXW8 A0A194Q5D3 A0A1B6FXI8 A0A0T6BBG6 A0A182RJN9 A0A0L7KQY3 A0A2A4K8M0 A0A139WK20 N6T731 A0A182Q4Y8 A0A182Y2E6 A0A182JHX3 A0A084WK02 A0A1B6L8W3 A0A182N976 J3JX86 A0A1S3CUY4 A0A1B6JLD2 E0W1R9 A0A067RLU5 A0A182VMN9 A0A182TV85 A0A182I4P2 Q5TQT2 A0A154PLK4 A0A0Q9XC88 B4KHL1 A0A182X4P5 A0A1I8N3X4 B7PN33 A0A182LK68 A0A182FDU9 A0A0L7R9E7 A0A1B6EAM9 A0A1W4V6M9 A0A2W1BL26 A0A0L8G521 A0A1Z5KVU7 H9JVW3 A0A232FJI8 A0A088A5Q7 V5HZ96 K7J083 B4N101 A0A023F6M6 A0A0K8W4C0 A0A0V0GB29 A0A1I8PB84 A0A1Q3FPV4 A0A034WIT6 B4M9B2 A0A1I8PQ47 A0A2A3E511 U5EQW5 B4JCS3 A0A151I4G8 A0A0J7KIH0 A0A131Z5C4 A0A151WQP9 A0A1S4EUT2 Q0C7B5 A0A195D0Q0 A0A0A1WSR5 A0A069DYI9 A0A182GUN5 B0WAN4 A0A023F8D4 A0A224XS15 A0A2P6KP25 A0A1I8MAA6 A0A2W1BSK7 Q9VQJ7 A0A0V0GCM6 B3NAN4 E1ZUW4 A0A0P5TK85 A0A2A4J5B2 A0A195FTI0 B4I2T4 B4NXB4 A0A0P5JYQ5 A0A0A9XWM0
Pubmed
19121390
26354079
26227816
23622113
28004739
18362917
+ More
19820115 23537049 25244985 24438588 22516182 20566863 24845553 12364791 14747013 17210077 17994087 25315136 20966253 28756777 28528879 28648823 25765539 20075255 25474469 25348373 18057021 26830274 17510324 25830018 26334808 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17550304 25401762
19820115 23537049 25244985 24438588 22516182 20566863 24845553 12364791 14747013 17210077 17994087 25315136 20966253 28756777 28528879 28648823 25765539 20075255 25474469 25348373 18057021 26830274 17510324 25830018 26334808 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17550304 25401762
EMBL
BABH01024121
NWSH01000747
PCG74438.1
ODYU01007653
SOQ50614.1
KQ459460
+ More
KPJ00769.1 JTDY01007828 KOB64914.1 GAIX01003850 JAA88710.1 KQ459838 KPJ19749.1 GEZM01024163 JAV87929.1 GAIX01004543 JAA88017.1 KQ971334 EEZ99699.1 KQ460141 KPJ17475.1 KPJ00753.1 GECZ01014862 JAS54907.1 LJIG01002295 KRT84645.1 JTDY01006795 KOB65683.1 NWSH01000066 PCG80012.1 KYB28263.1 APGK01041342 KB740993 KB632256 ENN76024.1 ERL90752.1 AXCN02000557 ATLV01024090 KE525349 KFB50546.1 GEBQ01019824 JAT20153.1 BT127855 AEE62817.1 GECU01007662 JAT00045.1 DS235873 EEB19651.1 KK852598 KDR20533.1 APCN01002341 AAAB01008960 EAL40132.3 KQ434972 KZC12749.1 CH933807 KRG03178.1 EDW12290.2 ABJB010351313 ABJB011001646 ABJB011007140 ABJB011048839 DS749781 EEC08005.1 KQ414627 KOC67478.1 GEDC01002364 JAS34934.1 KZ149984 PZC75739.1 KQ424055 KOF71670.1 GFJQ02007771 JAV99198.1 BABH01038693 NNAY01000123 OXU30753.1 GANP01004455 JAB80013.1 CH963920 EDW78163.1 GBBI01001830 JAC16882.1 GDHF01006368 JAI45946.1 GECL01000949 JAP05175.1 GFDL01005420 JAV29625.1 GAKP01005275 GAKP01005274 JAC53678.1 CH940654 EDW57788.1 KRF77953.1 KZ288400 PBC26259.1 GANO01004031 JAB55840.1 CH916368 EDW03162.1 KQ976448 KYM85872.1 LBMM01007086 KMQ90054.1 GEDV01002587 JAP85970.1 KQ982821 KYQ50184.1 CH477186 EAT48911.1 KQ977004 KYN06493.1 GBXI01012435 JAD01857.1 GBGD01002310 JAC86579.1 JXUM01000715 DS231873 EDS41588.1 GBBI01001116 JAC17596.1 GFTR01005104 JAW11322.1 MWRG01007752 PRD28085.1 PZC75740.1 AE014134 AY070563 AAF51172.1 AAL48034.1 GECL01000257 JAP05867.1 CH954177 EDV57557.1 GL434309 EFN75025.1 GDIP01126660 JAL77054.1 NWSH01002906 PCG67327.1 KQ981276 KYN43597.1 CH480820 EDW54079.1 CM000157 EDW87471.1 KRJ97169.1 GDIQ01191772 JAK59953.1 GBHO01019803 JAG23801.1
KPJ00769.1 JTDY01007828 KOB64914.1 GAIX01003850 JAA88710.1 KQ459838 KPJ19749.1 GEZM01024163 JAV87929.1 GAIX01004543 JAA88017.1 KQ971334 EEZ99699.1 KQ460141 KPJ17475.1 KPJ00753.1 GECZ01014862 JAS54907.1 LJIG01002295 KRT84645.1 JTDY01006795 KOB65683.1 NWSH01000066 PCG80012.1 KYB28263.1 APGK01041342 KB740993 KB632256 ENN76024.1 ERL90752.1 AXCN02000557 ATLV01024090 KE525349 KFB50546.1 GEBQ01019824 JAT20153.1 BT127855 AEE62817.1 GECU01007662 JAT00045.1 DS235873 EEB19651.1 KK852598 KDR20533.1 APCN01002341 AAAB01008960 EAL40132.3 KQ434972 KZC12749.1 CH933807 KRG03178.1 EDW12290.2 ABJB010351313 ABJB011001646 ABJB011007140 ABJB011048839 DS749781 EEC08005.1 KQ414627 KOC67478.1 GEDC01002364 JAS34934.1 KZ149984 PZC75739.1 KQ424055 KOF71670.1 GFJQ02007771 JAV99198.1 BABH01038693 NNAY01000123 OXU30753.1 GANP01004455 JAB80013.1 CH963920 EDW78163.1 GBBI01001830 JAC16882.1 GDHF01006368 JAI45946.1 GECL01000949 JAP05175.1 GFDL01005420 JAV29625.1 GAKP01005275 GAKP01005274 JAC53678.1 CH940654 EDW57788.1 KRF77953.1 KZ288400 PBC26259.1 GANO01004031 JAB55840.1 CH916368 EDW03162.1 KQ976448 KYM85872.1 LBMM01007086 KMQ90054.1 GEDV01002587 JAP85970.1 KQ982821 KYQ50184.1 CH477186 EAT48911.1 KQ977004 KYN06493.1 GBXI01012435 JAD01857.1 GBGD01002310 JAC86579.1 JXUM01000715 DS231873 EDS41588.1 GBBI01001116 JAC17596.1 GFTR01005104 JAW11322.1 MWRG01007752 PRD28085.1 PZC75740.1 AE014134 AY070563 AAF51172.1 AAL48034.1 GECL01000257 JAP05867.1 CH954177 EDV57557.1 GL434309 EFN75025.1 GDIP01126660 JAL77054.1 NWSH01002906 PCG67327.1 KQ981276 KYN43597.1 CH480820 EDW54079.1 CM000157 EDW87471.1 KRJ97169.1 GDIQ01191772 JAK59953.1 GBHO01019803 JAG23801.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000037510
UP000053240
UP000075885
+ More
UP000007266 UP000075901 UP000075900 UP000019118 UP000030742 UP000075886 UP000076408 UP000075880 UP000030765 UP000075884 UP000079169 UP000009046 UP000027135 UP000075903 UP000075902 UP000075840 UP000007062 UP000076502 UP000009192 UP000076407 UP000095301 UP000001555 UP000075882 UP000069272 UP000053825 UP000192221 UP000053454 UP000215335 UP000005203 UP000002358 UP000007798 UP000095300 UP000008792 UP000242457 UP000001070 UP000078540 UP000036403 UP000075809 UP000008820 UP000078542 UP000069940 UP000002320 UP000000803 UP000008711 UP000000311 UP000078541 UP000001292 UP000002282
UP000007266 UP000075901 UP000075900 UP000019118 UP000030742 UP000075886 UP000076408 UP000075880 UP000030765 UP000075884 UP000079169 UP000009046 UP000027135 UP000075903 UP000075902 UP000075840 UP000007062 UP000076502 UP000009192 UP000076407 UP000095301 UP000001555 UP000075882 UP000069272 UP000053825 UP000192221 UP000053454 UP000215335 UP000005203 UP000002358 UP000007798 UP000095300 UP000008792 UP000242457 UP000001070 UP000078540 UP000036403 UP000075809 UP000008820 UP000078542 UP000069940 UP000002320 UP000000803 UP000008711 UP000000311 UP000078541 UP000001292 UP000002282
Interpro
Gene 3D
ProteinModelPortal
H9IS98
A0A2A4JRW8
A0A2H1WDQ2
A0A194Q5X9
A0A0L7KPL8
S4PLU4
+ More
A0A0N0PES1 A0A1Y1MQH1 S4PXS2 A0A182PMH4 D6WI30 A0A0N1IIR4 A0A182SXW8 A0A194Q5D3 A0A1B6FXI8 A0A0T6BBG6 A0A182RJN9 A0A0L7KQY3 A0A2A4K8M0 A0A139WK20 N6T731 A0A182Q4Y8 A0A182Y2E6 A0A182JHX3 A0A084WK02 A0A1B6L8W3 A0A182N976 J3JX86 A0A1S3CUY4 A0A1B6JLD2 E0W1R9 A0A067RLU5 A0A182VMN9 A0A182TV85 A0A182I4P2 Q5TQT2 A0A154PLK4 A0A0Q9XC88 B4KHL1 A0A182X4P5 A0A1I8N3X4 B7PN33 A0A182LK68 A0A182FDU9 A0A0L7R9E7 A0A1B6EAM9 A0A1W4V6M9 A0A2W1BL26 A0A0L8G521 A0A1Z5KVU7 H9JVW3 A0A232FJI8 A0A088A5Q7 V5HZ96 K7J083 B4N101 A0A023F6M6 A0A0K8W4C0 A0A0V0GB29 A0A1I8PB84 A0A1Q3FPV4 A0A034WIT6 B4M9B2 A0A1I8PQ47 A0A2A3E511 U5EQW5 B4JCS3 A0A151I4G8 A0A0J7KIH0 A0A131Z5C4 A0A151WQP9 A0A1S4EUT2 Q0C7B5 A0A195D0Q0 A0A0A1WSR5 A0A069DYI9 A0A182GUN5 B0WAN4 A0A023F8D4 A0A224XS15 A0A2P6KP25 A0A1I8MAA6 A0A2W1BSK7 Q9VQJ7 A0A0V0GCM6 B3NAN4 E1ZUW4 A0A0P5TK85 A0A2A4J5B2 A0A195FTI0 B4I2T4 B4NXB4 A0A0P5JYQ5 A0A0A9XWM0
A0A0N0PES1 A0A1Y1MQH1 S4PXS2 A0A182PMH4 D6WI30 A0A0N1IIR4 A0A182SXW8 A0A194Q5D3 A0A1B6FXI8 A0A0T6BBG6 A0A182RJN9 A0A0L7KQY3 A0A2A4K8M0 A0A139WK20 N6T731 A0A182Q4Y8 A0A182Y2E6 A0A182JHX3 A0A084WK02 A0A1B6L8W3 A0A182N976 J3JX86 A0A1S3CUY4 A0A1B6JLD2 E0W1R9 A0A067RLU5 A0A182VMN9 A0A182TV85 A0A182I4P2 Q5TQT2 A0A154PLK4 A0A0Q9XC88 B4KHL1 A0A182X4P5 A0A1I8N3X4 B7PN33 A0A182LK68 A0A182FDU9 A0A0L7R9E7 A0A1B6EAM9 A0A1W4V6M9 A0A2W1BL26 A0A0L8G521 A0A1Z5KVU7 H9JVW3 A0A232FJI8 A0A088A5Q7 V5HZ96 K7J083 B4N101 A0A023F6M6 A0A0K8W4C0 A0A0V0GB29 A0A1I8PB84 A0A1Q3FPV4 A0A034WIT6 B4M9B2 A0A1I8PQ47 A0A2A3E511 U5EQW5 B4JCS3 A0A151I4G8 A0A0J7KIH0 A0A131Z5C4 A0A151WQP9 A0A1S4EUT2 Q0C7B5 A0A195D0Q0 A0A0A1WSR5 A0A069DYI9 A0A182GUN5 B0WAN4 A0A023F8D4 A0A224XS15 A0A2P6KP25 A0A1I8MAA6 A0A2W1BSK7 Q9VQJ7 A0A0V0GCM6 B3NAN4 E1ZUW4 A0A0P5TK85 A0A2A4J5B2 A0A195FTI0 B4I2T4 B4NXB4 A0A0P5JYQ5 A0A0A9XWM0
PDB
1Y97
E-value=5.44016e-16,
Score=204
Ontologies
GO
PANTHER
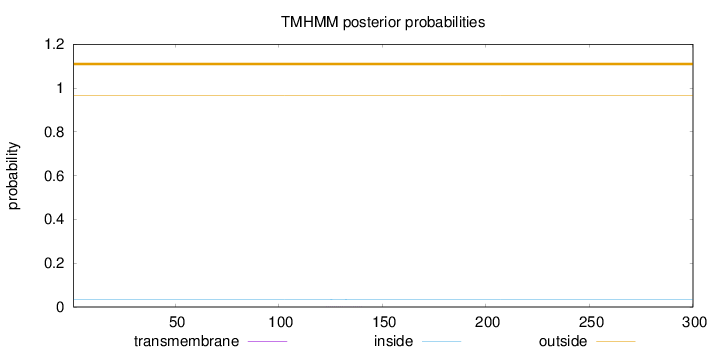
Topology
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00547
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03471
outside
1 - 300
Population Genetic Test Statistics
Pi
189.6139
Theta
163.845642
Tajima's D
0.31816
CLR
448.145007
CSRT
0.454427278636068
Interpretation
Uncertain
