Gene
KWMTBOMO14418
Pre Gene Modal
BGIBMGA000123
Annotation
PREDICTED:_desumoylating_isopeptidase_2_isoform_X4_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.702
Sequence
CDS
ATGTCGCTGCTATCGCGCCGTTCAGACTCCCAGCACCCTCGGCCCGGACAGGCGACCGTGGTCCTCAACGTTTACGATATGTATTGGACGAATTGGTACACTGCGGGCGCGGGTCTGGGTGTGTTCCACAGCGGTGTTCAAGTTCACGGATCGGAATGGGCGTACGGGGGTCATCCGTACGCGTTCACCGGGGTCTTCGAGATATCGCCGCGAGACGAACGGGAACTAGGGGAGCAGTTTCGATTCAGGCAGAGTGTACATATAGGATATACAGATTTCAGCGAGGAGGAAGTAAGAAGATTAGTCGCAGAACTAGGGAAGCAATTCAGAGGTGACAGATACCATTTAATGAACAATAACTGCAATCATTTCACTTCAGCCTTTTGCATGGCGTTGTGCTCACGCGACATTCCGCCGTGGGTGAATCGGCTAGCGTACGTGAGCTCGTGCGTTCCATTCCTGCAGAGGTGTCTACCGAAGGAGTGGCTGACGCCGGCGGCCCTGCAGCAGTCGTTGGCAGCGCACTCGCCATCCTCCTCTTCGCAGCACTCTCCCTAA
Protein
MSLLSRRSDSQHPRPGQATVVLNVYDMYWTNWYTAGAGLGVFHSGVQVHGSEWAYGGHPYAFTGVFEISPRDERELGEQFRFRQSVHIGYTDFSEEEVRRLVAELGKQFRGDRYHLMNNNCNHFTSAFCMALCSRDIPPWVNRLAYVSSCVPFLQRCLPKEWLTPAALQQSLAAHSPSSSSQHSP
Summary
Uniprot
A0A2A4JVV3
A0A2H1VH45
A0A194RII3
A0A2P8Y0J6
A0A1W4WUA7
A0A2J7QV36
+ More
A0A067RAG1 U5EQ18 D6WG68 A0A1B0CCZ0 A0A1L8DTS9 A0A1B6C1N8 J3JUN5 A0A232FJE7 K7IZC1 A0A1B6CJR0 A0A0K8TSN7 A0A0L7QMF2 A0A151HYS4 A0A158NSR1 A0A1I8MEX6 A0A0L0CQR2 A0A0K8U3M0 A0A0C9R3K9 A0A0P4W1K4 W8C6W2 A0A034WTS7 A0A0C9RA09 A0A0A1WW17 V5GL94 A0A1B6FZ32 A0A1B6M3Y7 A0A1B6H9W6 S6CVW6 A0A1A9ZRC8 A0A1A9WAD0 A0A2P2HYX2 A0A1B0FL39 A0A1I8PAE5 A0A154NZ37 E9IN41 A0A026WID4 A0A0M9A140 A0A1B6K2P9 A0A1B6FAP7 A0A195CD52 F4W879 A0A151X006 B4J9E7 A0A1Y1MJK2 A0A195DV04 A0A1B6KFU1 A0A2A3EAI0 A0A1Y1MMG0 V9ID43 A0A088ALG9 A0A1W4VRR8 A0A3B0JLN7 Q28XK2 B4GG98 B4P831 B4HN05 B3NN48 B4QAN3 Q7K2B1 B4KUC6 B4LPI1 A0A195EWW7 A0A1J1IID5 B3MIK7 B4MK70 A0A0M5IX37 D3TPM2 E9GIU5 A0A2T7NW57 A0A0P6DBE4 A0A023FAC9 C3Y071 A0A224XT21 A0A0V0GCG1 A0A069DQN7 A0A1S3D5V5 A0A131XU09 A0A0A9XLE5 A0A146LLB7 B7QAI7 A0A0K8SSH8 V5H6W8 A0A0N7Z9I1 T1HZK7 A0A2C9JEX3 A0A0A9XGL1 A0A146KQH8 C1BQI7 A0A1S4F664 A0A182X096 A0A1D2N6S1 A0A182LUV7
A0A067RAG1 U5EQ18 D6WG68 A0A1B0CCZ0 A0A1L8DTS9 A0A1B6C1N8 J3JUN5 A0A232FJE7 K7IZC1 A0A1B6CJR0 A0A0K8TSN7 A0A0L7QMF2 A0A151HYS4 A0A158NSR1 A0A1I8MEX6 A0A0L0CQR2 A0A0K8U3M0 A0A0C9R3K9 A0A0P4W1K4 W8C6W2 A0A034WTS7 A0A0C9RA09 A0A0A1WW17 V5GL94 A0A1B6FZ32 A0A1B6M3Y7 A0A1B6H9W6 S6CVW6 A0A1A9ZRC8 A0A1A9WAD0 A0A2P2HYX2 A0A1B0FL39 A0A1I8PAE5 A0A154NZ37 E9IN41 A0A026WID4 A0A0M9A140 A0A1B6K2P9 A0A1B6FAP7 A0A195CD52 F4W879 A0A151X006 B4J9E7 A0A1Y1MJK2 A0A195DV04 A0A1B6KFU1 A0A2A3EAI0 A0A1Y1MMG0 V9ID43 A0A088ALG9 A0A1W4VRR8 A0A3B0JLN7 Q28XK2 B4GG98 B4P831 B4HN05 B3NN48 B4QAN3 Q7K2B1 B4KUC6 B4LPI1 A0A195EWW7 A0A1J1IID5 B3MIK7 B4MK70 A0A0M5IX37 D3TPM2 E9GIU5 A0A2T7NW57 A0A0P6DBE4 A0A023FAC9 C3Y071 A0A224XT21 A0A0V0GCG1 A0A069DQN7 A0A1S3D5V5 A0A131XU09 A0A0A9XLE5 A0A146LLB7 B7QAI7 A0A0K8SSH8 V5H6W8 A0A0N7Z9I1 T1HZK7 A0A2C9JEX3 A0A0A9XGL1 A0A146KQH8 C1BQI7 A0A1S4F664 A0A182X096 A0A1D2N6S1 A0A182LUV7
Pubmed
26354079
29403074
24845553
18362917
19820115
22516182
+ More
23537049 28648823 20075255 26369729 21347285 25315136 26108605 24495485 25348373 25830018 23938757 21282665 24508170 30249741 21719571 17994087 28004739 15632085 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 20353571 21292972 25474469 18563158 26334808 25401762 26823975 25765539 27129103 15562597 27289101
23537049 28648823 20075255 26369729 21347285 25315136 26108605 24495485 25348373 25830018 23938757 21282665 24508170 30249741 21719571 17994087 28004739 15632085 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 20353571 21292972 25474469 18563158 26334808 25401762 26823975 25765539 27129103 15562597 27289101
EMBL
NWSH01000581
PCG75520.1
ODYU01002345
SOQ39722.1
KQ460124
KPJ17633.1
+ More
PYGN01001080 PSN37788.1 NEVH01010479 PNF32449.1 KK852595 KDR20630.1 GANO01003467 JAB56404.1 KQ971320 EFA00538.1 AJWK01007243 GFDF01004310 JAV09774.1 GEDC01030014 GEDC01023785 JAS07284.1 JAS13513.1 APGK01021662 BT126950 KB740350 KB632240 AEE61912.1 ENN80847.1 ERL90531.1 NNAY01000112 OXU30864.1 GEDC01023656 JAS13642.1 GDAI01000465 JAI17138.1 KQ414889 KOC59823.1 KQ976723 KYM76678.1 ADTU01025113 JRES01000153 KNC33774.1 GDHF01031032 JAI21282.1 GBYB01010904 JAG80671.1 GDRN01099818 JAI58700.1 GAMC01000902 JAC05654.1 GAKP01001774 JAC57178.1 GBYB01009812 JAG79579.1 GBXI01011511 JAD02781.1 GALX01006089 JAB62377.1 GECZ01014321 JAS55448.1 GEBQ01009354 JAT30623.1 GECU01036255 JAS71451.1 HF586479 CCQ71376.1 IACF01001260 LAB66972.1 CCAG010020342 KQ434778 KZC04324.1 GL764272 EFZ18015.1 KK107191 QOIP01000009 EZA55728.1 RLU18554.1 KQ435794 KOX73723.1 GECU01001998 JAT05709.1 GECZ01022565 JAS47204.1 KQ978023 KYM98008.1 GL887898 EGI69574.1 KQ982624 KYQ53684.1 CH916367 EDW01428.1 GEZM01029913 JAV85884.1 KQ980322 KYN16577.1 GEBQ01029649 JAT10328.1 KZ288324 PBC28071.1 GEZM01029915 JAV85880.1 JR038391 JR038392 AEY58233.1 AEY58234.1 OUUW01000001 SPP74156.1 CM000071 EAL26314.1 CH479183 EDW35518.1 CM000158 EDW90078.1 CH480816 EDW47309.1 CH954179 EDV56568.1 CM000362 CM002911 EDX06538.1 KMY92856.1 AE013599 AY061130 AAF58750.1 AAL28678.1 CH933808 EDW09722.1 KRG04864.1 CH940648 EDW61240.1 KQ981948 KYN32646.1 CVRI01000054 CRL00005.1 CH902619 EDV38083.1 CH963846 EDW72509.1 CP012524 ALC40674.1 EZ423374 ADD19650.1 GL732547 EFX80329.1 PZQS01000008 PVD25408.1 GDIP01090268 GDIQ01080567 LRGB01002993 JAM13447.1 JAN14170.1 KZS05192.1 GBBI01000824 JAC17888.1 GG666478 EEN66144.1 GFTR01003458 JAW12968.1 GECL01000676 JAP05448.1 GBGD01002952 JAC85937.1 GEFM01006111 JAP69685.1 GBHO01025657 GBHO01025654 GBHO01025652 JAG17947.1 JAG17950.1 JAG17952.1 GDHC01011092 GDHC01009700 JAQ07537.1 JAQ08929.1 ABJB010700554 ABJB011104654 DS895249 EEC15859.1 GBRD01009507 JAG56317.1 GANP01011684 JAB72784.1 GDKW01000483 JAI56112.1 ACPB03007516 GBHO01025656 JAG17948.1 GDHC01019856 JAP98772.1 BT076866 ACO11290.1 LJIJ01000179 ODN00954.1 AXCM01005558
PYGN01001080 PSN37788.1 NEVH01010479 PNF32449.1 KK852595 KDR20630.1 GANO01003467 JAB56404.1 KQ971320 EFA00538.1 AJWK01007243 GFDF01004310 JAV09774.1 GEDC01030014 GEDC01023785 JAS07284.1 JAS13513.1 APGK01021662 BT126950 KB740350 KB632240 AEE61912.1 ENN80847.1 ERL90531.1 NNAY01000112 OXU30864.1 GEDC01023656 JAS13642.1 GDAI01000465 JAI17138.1 KQ414889 KOC59823.1 KQ976723 KYM76678.1 ADTU01025113 JRES01000153 KNC33774.1 GDHF01031032 JAI21282.1 GBYB01010904 JAG80671.1 GDRN01099818 JAI58700.1 GAMC01000902 JAC05654.1 GAKP01001774 JAC57178.1 GBYB01009812 JAG79579.1 GBXI01011511 JAD02781.1 GALX01006089 JAB62377.1 GECZ01014321 JAS55448.1 GEBQ01009354 JAT30623.1 GECU01036255 JAS71451.1 HF586479 CCQ71376.1 IACF01001260 LAB66972.1 CCAG010020342 KQ434778 KZC04324.1 GL764272 EFZ18015.1 KK107191 QOIP01000009 EZA55728.1 RLU18554.1 KQ435794 KOX73723.1 GECU01001998 JAT05709.1 GECZ01022565 JAS47204.1 KQ978023 KYM98008.1 GL887898 EGI69574.1 KQ982624 KYQ53684.1 CH916367 EDW01428.1 GEZM01029913 JAV85884.1 KQ980322 KYN16577.1 GEBQ01029649 JAT10328.1 KZ288324 PBC28071.1 GEZM01029915 JAV85880.1 JR038391 JR038392 AEY58233.1 AEY58234.1 OUUW01000001 SPP74156.1 CM000071 EAL26314.1 CH479183 EDW35518.1 CM000158 EDW90078.1 CH480816 EDW47309.1 CH954179 EDV56568.1 CM000362 CM002911 EDX06538.1 KMY92856.1 AE013599 AY061130 AAF58750.1 AAL28678.1 CH933808 EDW09722.1 KRG04864.1 CH940648 EDW61240.1 KQ981948 KYN32646.1 CVRI01000054 CRL00005.1 CH902619 EDV38083.1 CH963846 EDW72509.1 CP012524 ALC40674.1 EZ423374 ADD19650.1 GL732547 EFX80329.1 PZQS01000008 PVD25408.1 GDIP01090268 GDIQ01080567 LRGB01002993 JAM13447.1 JAN14170.1 KZS05192.1 GBBI01000824 JAC17888.1 GG666478 EEN66144.1 GFTR01003458 JAW12968.1 GECL01000676 JAP05448.1 GBGD01002952 JAC85937.1 GEFM01006111 JAP69685.1 GBHO01025657 GBHO01025654 GBHO01025652 JAG17947.1 JAG17950.1 JAG17952.1 GDHC01011092 GDHC01009700 JAQ07537.1 JAQ08929.1 ABJB010700554 ABJB011104654 DS895249 EEC15859.1 GBRD01009507 JAG56317.1 GANP01011684 JAB72784.1 GDKW01000483 JAI56112.1 ACPB03007516 GBHO01025656 JAG17948.1 GDHC01019856 JAP98772.1 BT076866 ACO11290.1 LJIJ01000179 ODN00954.1 AXCM01005558
Proteomes
UP000218220
UP000053240
UP000245037
UP000192223
UP000235965
UP000027135
+ More
UP000007266 UP000092461 UP000019118 UP000030742 UP000215335 UP000002358 UP000053825 UP000078540 UP000005205 UP000095301 UP000037069 UP000092445 UP000091820 UP000092444 UP000095300 UP000076502 UP000053097 UP000279307 UP000053105 UP000078542 UP000007755 UP000075809 UP000001070 UP000078492 UP000242457 UP000005203 UP000192221 UP000268350 UP000001819 UP000008744 UP000002282 UP000001292 UP000008711 UP000000304 UP000000803 UP000009192 UP000008792 UP000078541 UP000183832 UP000007801 UP000007798 UP000092553 UP000000305 UP000245119 UP000076858 UP000001554 UP000079169 UP000001555 UP000015103 UP000076420 UP000076407 UP000094527 UP000075883
UP000007266 UP000092461 UP000019118 UP000030742 UP000215335 UP000002358 UP000053825 UP000078540 UP000005205 UP000095301 UP000037069 UP000092445 UP000091820 UP000092444 UP000095300 UP000076502 UP000053097 UP000279307 UP000053105 UP000078542 UP000007755 UP000075809 UP000001070 UP000078492 UP000242457 UP000005203 UP000192221 UP000268350 UP000001819 UP000008744 UP000002282 UP000001292 UP000008711 UP000000304 UP000000803 UP000009192 UP000008792 UP000078541 UP000183832 UP000007801 UP000007798 UP000092553 UP000000305 UP000245119 UP000076858 UP000001554 UP000079169 UP000001555 UP000015103 UP000076420 UP000076407 UP000094527 UP000075883
PRIDE
Pfam
PF05903 Peptidase_C97
Gene 3D
ProteinModelPortal
A0A2A4JVV3
A0A2H1VH45
A0A194RII3
A0A2P8Y0J6
A0A1W4WUA7
A0A2J7QV36
+ More
A0A067RAG1 U5EQ18 D6WG68 A0A1B0CCZ0 A0A1L8DTS9 A0A1B6C1N8 J3JUN5 A0A232FJE7 K7IZC1 A0A1B6CJR0 A0A0K8TSN7 A0A0L7QMF2 A0A151HYS4 A0A158NSR1 A0A1I8MEX6 A0A0L0CQR2 A0A0K8U3M0 A0A0C9R3K9 A0A0P4W1K4 W8C6W2 A0A034WTS7 A0A0C9RA09 A0A0A1WW17 V5GL94 A0A1B6FZ32 A0A1B6M3Y7 A0A1B6H9W6 S6CVW6 A0A1A9ZRC8 A0A1A9WAD0 A0A2P2HYX2 A0A1B0FL39 A0A1I8PAE5 A0A154NZ37 E9IN41 A0A026WID4 A0A0M9A140 A0A1B6K2P9 A0A1B6FAP7 A0A195CD52 F4W879 A0A151X006 B4J9E7 A0A1Y1MJK2 A0A195DV04 A0A1B6KFU1 A0A2A3EAI0 A0A1Y1MMG0 V9ID43 A0A088ALG9 A0A1W4VRR8 A0A3B0JLN7 Q28XK2 B4GG98 B4P831 B4HN05 B3NN48 B4QAN3 Q7K2B1 B4KUC6 B4LPI1 A0A195EWW7 A0A1J1IID5 B3MIK7 B4MK70 A0A0M5IX37 D3TPM2 E9GIU5 A0A2T7NW57 A0A0P6DBE4 A0A023FAC9 C3Y071 A0A224XT21 A0A0V0GCG1 A0A069DQN7 A0A1S3D5V5 A0A131XU09 A0A0A9XLE5 A0A146LLB7 B7QAI7 A0A0K8SSH8 V5H6W8 A0A0N7Z9I1 T1HZK7 A0A2C9JEX3 A0A0A9XGL1 A0A146KQH8 C1BQI7 A0A1S4F664 A0A182X096 A0A1D2N6S1 A0A182LUV7
A0A067RAG1 U5EQ18 D6WG68 A0A1B0CCZ0 A0A1L8DTS9 A0A1B6C1N8 J3JUN5 A0A232FJE7 K7IZC1 A0A1B6CJR0 A0A0K8TSN7 A0A0L7QMF2 A0A151HYS4 A0A158NSR1 A0A1I8MEX6 A0A0L0CQR2 A0A0K8U3M0 A0A0C9R3K9 A0A0P4W1K4 W8C6W2 A0A034WTS7 A0A0C9RA09 A0A0A1WW17 V5GL94 A0A1B6FZ32 A0A1B6M3Y7 A0A1B6H9W6 S6CVW6 A0A1A9ZRC8 A0A1A9WAD0 A0A2P2HYX2 A0A1B0FL39 A0A1I8PAE5 A0A154NZ37 E9IN41 A0A026WID4 A0A0M9A140 A0A1B6K2P9 A0A1B6FAP7 A0A195CD52 F4W879 A0A151X006 B4J9E7 A0A1Y1MJK2 A0A195DV04 A0A1B6KFU1 A0A2A3EAI0 A0A1Y1MMG0 V9ID43 A0A088ALG9 A0A1W4VRR8 A0A3B0JLN7 Q28XK2 B4GG98 B4P831 B4HN05 B3NN48 B4QAN3 Q7K2B1 B4KUC6 B4LPI1 A0A195EWW7 A0A1J1IID5 B3MIK7 B4MK70 A0A0M5IX37 D3TPM2 E9GIU5 A0A2T7NW57 A0A0P6DBE4 A0A023FAC9 C3Y071 A0A224XT21 A0A0V0GCG1 A0A069DQN7 A0A1S3D5V5 A0A131XU09 A0A0A9XLE5 A0A146LLB7 B7QAI7 A0A0K8SSH8 V5H6W8 A0A0N7Z9I1 T1HZK7 A0A2C9JEX3 A0A0A9XGL1 A0A146KQH8 C1BQI7 A0A1S4F664 A0A182X096 A0A1D2N6S1 A0A182LUV7
PDB
3EBQ
E-value=9.89681e-07,
Score=122
Ontologies
GO
PANTHER
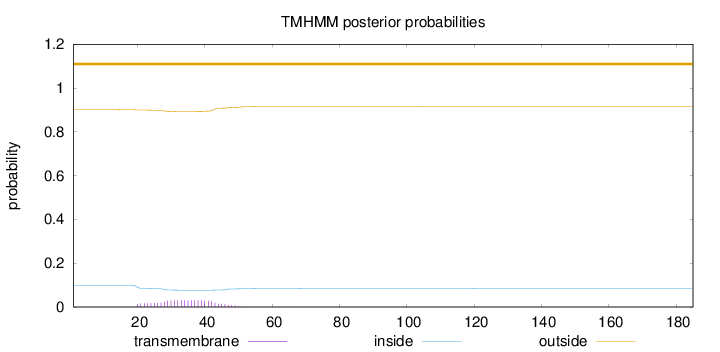
Topology
Length:
185
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.676819999999999
Exp number, first 60 AAs:
0.67115
Total prob of N-in:
0.09940
outside
1 - 185
Population Genetic Test Statistics
Pi
193.871062
Theta
159.488887
Tajima's D
0.58936
CLR
1.589684
CSRT
0.539173041347933
Interpretation
Uncertain
