Gene
KWMTBOMO14414 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000106
Annotation
mitochondrial_intermembrane_space_translocase_subunit_Tim10_[Bombyx_mori]
Full name
Mitochondrial import inner membrane translocase subunit Tim10
Location in the cell
Nuclear Reliability : 2.326
Sequence
CDS
ATGGCAGTACCGCAGCTAGACCCTGCAAAGCTGCAATTGGTGCAGGAACTGGAGATTGAGATGATGTCCGACATGTACAACCGCTTGGTGAGCGCATGTCACCGGAAGTGCATACCAATTAAATATCATGAACCTGAATTAGGAAAAGGCGAATCTGTATGTTTAGATCGCTGTGTAGCCAAATACTTGGACGTACATGAACGCATAGGTAAGAAACTTTCCAATATGTCACAAGGAGAAACCGAAGATCTCACAAAGGTCAACATACCAGACAAAAAATAG
Protein
MAVPQLDPAKLQLVQELEIEMMSDMYNRLVSACHRKCIPIKYHEPELGKGESVCLDRCVAKYLDVHERIGKKLSNMSQGETEDLTKVNIPDKK
Summary
Description
Mitochondrial intermembrane chaperone that participates in the import and insertion of multi-pass transmembrane proteins into the mitochondrial inner membrane. May also be required for the transfer of beta-barrel precursors from the TOM complex to the sorting and assembly machinery (SAM complex) of the outer membrane. Acts as a chaperone-like protein that protects the hydrophobic precursors from aggregation and guide them through the mitochondrial intermembrane space (By similarity).
Subunit
Heterohexamer; composed of 3 copies of Tim9 and 3 copies of Tim10, named soluble 70 kDa complex. The complex associates with the Tim22 component of the TIM22 complex. Interacts with multi-pass transmembrane proteins in transit (By similarity).
Similarity
Belongs to the small Tim family.
Keywords
Chaperone
Complete proteome
Disulfide bond
Membrane
Metal-binding
Mitochondrion
Mitochondrion inner membrane
Protein transport
Reference proteome
Translocation
Transport
Zinc
Feature
chain Mitochondrial import inner membrane translocase subunit Tim10
Uniprot
Q2F5K3
A0A3S2LV35
I4DJZ5
A0A194RJV3
A0A212FA50
A0A194QH60
+ More
A0A2A4JV82 A0A2H1VDU2 Q9Y0V4 E2AQP5 A0A087U6B3 A0A067QQ30 A0A2R5LHJ0 R4V508 A0A195D9W6 E0VY47 A0A195F540 A0A151IEZ5 F4W525 A0A195BQ48 A0A158NDJ4 A0A026W2F0 E2B619 A0A023G8F3 E9GAU9 A0A023FR62 A0A0K8R6Y7 A0A1Z5LE75 A0A151XEG9 A0A1E1XV82 A0A0P5ELG7 A0A023FP94 A0A023FVH6 A0A1E1X0C6 A0A131Z5G8 A0A224Z4J2 L7LXC6 A0A131XE60 A0A2J7QM11 A0A0C9QYR7 A0A182U576 U5EZ06 A0A293MEQ1 Q7QE31 A0A1B2AKF1 B4I7U7 B4QGJ5 Q9W2D6 B3MIZ0 T1DHV3 A0A0Q9WYL8 A0A1J1HX77 A0A1B0ARR9 K7IMJ3 A0A3S3NJ03 A0A0M3QV06 A0A182WY03 B4LNP8 A0A1A9WV14 B4JVF8 Q1HQZ1 A0A023EDY9 E9J336 A0A154PHR5 A0A2M3Z7I4 A0A2M4ATZ9 W5JQC8 A0A1B0FD43 A0A2R7W3S0 A0A023EB85 A0A182FVZ1 B4KTP5 A0A1Q3FD67 B0WTF8 A0A1A9ZR57 A0A182JQ62 A0A084WUG2 A0A182I623 A0A0U2TLH7 A0A182YAB3 A0A182PN56 A0A0P4VQ74 R4G4L5 A0A182LZ46 T1E892 A0A023F9G4 C1BP39 A0A224XUT1 C1BRK1 A0A0V0G8G8 A0A069DNN5 A0A0K2U8Z5 A0A182RLV6 A0A336LD03 A0A182NP99 A0A3B0JP97 Q292C1 B4G9X1 C1C134 A0A182WE87
A0A2A4JV82 A0A2H1VDU2 Q9Y0V4 E2AQP5 A0A087U6B3 A0A067QQ30 A0A2R5LHJ0 R4V508 A0A195D9W6 E0VY47 A0A195F540 A0A151IEZ5 F4W525 A0A195BQ48 A0A158NDJ4 A0A026W2F0 E2B619 A0A023G8F3 E9GAU9 A0A023FR62 A0A0K8R6Y7 A0A1Z5LE75 A0A151XEG9 A0A1E1XV82 A0A0P5ELG7 A0A023FP94 A0A023FVH6 A0A1E1X0C6 A0A131Z5G8 A0A224Z4J2 L7LXC6 A0A131XE60 A0A2J7QM11 A0A0C9QYR7 A0A182U576 U5EZ06 A0A293MEQ1 Q7QE31 A0A1B2AKF1 B4I7U7 B4QGJ5 Q9W2D6 B3MIZ0 T1DHV3 A0A0Q9WYL8 A0A1J1HX77 A0A1B0ARR9 K7IMJ3 A0A3S3NJ03 A0A0M3QV06 A0A182WY03 B4LNP8 A0A1A9WV14 B4JVF8 Q1HQZ1 A0A023EDY9 E9J336 A0A154PHR5 A0A2M3Z7I4 A0A2M4ATZ9 W5JQC8 A0A1B0FD43 A0A2R7W3S0 A0A023EB85 A0A182FVZ1 B4KTP5 A0A1Q3FD67 B0WTF8 A0A1A9ZR57 A0A182JQ62 A0A084WUG2 A0A182I623 A0A0U2TLH7 A0A182YAB3 A0A182PN56 A0A0P4VQ74 R4G4L5 A0A182LZ46 T1E892 A0A023F9G4 C1BP39 A0A224XUT1 C1BRK1 A0A0V0G8G8 A0A069DNN5 A0A0K2U8Z5 A0A182RLV6 A0A336LD03 A0A182NP99 A0A3B0JP97 Q292C1 B4G9X1 C1C134 A0A182WE87
Pubmed
19121390
22651552
26354079
22118469
20798317
24845553
+ More
20566863 21719571 21347285 24508170 30249741 21292972 28528879 29209593 28503490 26830274 28797301 25576852 28049606 26131772 12364791 14747013 17210077 17994087 22936249 10611480 10731132 12537572 12537569 24330624 20075255 17204158 17510324 24945155 26483478 21282665 20920257 23761445 24438588 26621068 25244985 27129103 25474469 26334808 15632085
20566863 21719571 21347285 24508170 30249741 21292972 28528879 29209593 28503490 26830274 28797301 25576852 28049606 26131772 12364791 14747013 17210077 17994087 22936249 10611480 10731132 12537572 12537569 24330624 20075255 17204158 17510324 24945155 26483478 21282665 20920257 23761445 24438588 26621068 25244985 27129103 25474469 26334808 15632085
EMBL
BABH01024106
DQ311420
ABD36364.1
RSAL01000179
RVE44972.1
AK401613
+ More
BAM18235.1 KQ460124 KPJ17615.1 AGBW02009528 OWR50589.1 KQ458859 KPJ04847.1 NWSH01000581 PCG75524.1 ODYU01002020 SOQ39008.1 AF150098 AAD40004.1 GL441804 EFN64234.1 KK118409 KFM72902.1 KK853149 KDR10665.1 GGLE01004874 MBY09000.1 KC741124 AGM32948.1 KQ981082 KYN09661.1 DS235843 EEB18303.1 KQ981820 KYN35189.1 KQ977830 KYM99400.1 GL887596 EGI70721.1 KQ976424 KYM88263.1 ADTU01012636 KK107474 QOIP01000002 EZA50173.1 RLU26109.1 GL445900 EFN88880.1 GBBM01006290 JAC29128.1 GL732537 EFX83320.1 GBBK01001372 JAC23110.1 GADI01007564 JAA66244.1 GFJQ02001729 JAW05241.1 KQ982254 KYQ58737.1 GFAA01000465 JAU02970.1 GDIP01144763 LRGB01000389 JAJ78639.1 KZS19400.1 GBBK01001373 JAC23109.1 GBBL01002562 JAC24758.1 GFAC01006463 JAT92725.1 GEDV01002552 JAP86005.1 GFPF01009814 MAA20960.1 GACK01008318 JAA56716.1 GEFH01004680 JAP63901.1 NEVH01013216 PNF29649.1 GBZX01002825 JAG89915.1 GANO01001655 JAB58216.1 GFWV01019469 MAA44197.1 AAAB01008848 EAA06991.2 KX531842 ANY27652.1 CH480824 EDW56672.1 CM000362 CM002911 EDX08114.1 KMY95624.1 AF150092 AE013599 AY061513 CH902619 EDV37056.1 GALA01001311 JAA93541.1 CH963719 KRF97484.1 CVRI01000033 CRK92629.1 JXJN01002564 NCKU01011698 NCKU01011636 RWS00346.1 RWS00376.1 CP012524 ALC41579.1 CH940648 EDW60118.1 CH916375 EDV98426.1 DQ440303 CH477663 ABF18336.1 EAT37572.1 JXUM01241297 GAPW01006592 KQ635200 JAC07006.1 KXJ62521.1 GL768055 EFZ12794.1 KQ434899 KZC11034.1 GGFM01003726 MBW24477.1 GGFK01010926 MBW44247.1 ADMH02000766 ETN65110.1 CCAG010002640 KK854300 PTY14364.1 GAPW01006721 JAC06877.1 CH933808 EDW09628.1 GFDL01009529 JAV25516.1 DS232085 EDS34396.1 ATLV01027083 KE525423 KFB53856.1 APCN01003547 KT755167 ALS05001.1 GDKW01001976 JAI54619.1 ACPB03008858 GAHY01001255 JAA76255.1 AXCM01001000 GAMD01002973 JAA98617.1 GBBI01001073 JAC17639.1 BT076368 ACO10792.1 GFTR01001512 JAW14914.1 BT077230 ACO11654.1 GECL01001870 JAP04254.1 GBGD01003618 JAC85271.1 HACA01017343 CDW34704.1 UFQS01002647 UFQT01002647 SSX14435.1 SSX33842.1 OUUW01000001 SPP75429.1 CM000071 EAL24941.1 CH479181 EDW31723.1 BT080563 ACO14987.1
BAM18235.1 KQ460124 KPJ17615.1 AGBW02009528 OWR50589.1 KQ458859 KPJ04847.1 NWSH01000581 PCG75524.1 ODYU01002020 SOQ39008.1 AF150098 AAD40004.1 GL441804 EFN64234.1 KK118409 KFM72902.1 KK853149 KDR10665.1 GGLE01004874 MBY09000.1 KC741124 AGM32948.1 KQ981082 KYN09661.1 DS235843 EEB18303.1 KQ981820 KYN35189.1 KQ977830 KYM99400.1 GL887596 EGI70721.1 KQ976424 KYM88263.1 ADTU01012636 KK107474 QOIP01000002 EZA50173.1 RLU26109.1 GL445900 EFN88880.1 GBBM01006290 JAC29128.1 GL732537 EFX83320.1 GBBK01001372 JAC23110.1 GADI01007564 JAA66244.1 GFJQ02001729 JAW05241.1 KQ982254 KYQ58737.1 GFAA01000465 JAU02970.1 GDIP01144763 LRGB01000389 JAJ78639.1 KZS19400.1 GBBK01001373 JAC23109.1 GBBL01002562 JAC24758.1 GFAC01006463 JAT92725.1 GEDV01002552 JAP86005.1 GFPF01009814 MAA20960.1 GACK01008318 JAA56716.1 GEFH01004680 JAP63901.1 NEVH01013216 PNF29649.1 GBZX01002825 JAG89915.1 GANO01001655 JAB58216.1 GFWV01019469 MAA44197.1 AAAB01008848 EAA06991.2 KX531842 ANY27652.1 CH480824 EDW56672.1 CM000362 CM002911 EDX08114.1 KMY95624.1 AF150092 AE013599 AY061513 CH902619 EDV37056.1 GALA01001311 JAA93541.1 CH963719 KRF97484.1 CVRI01000033 CRK92629.1 JXJN01002564 NCKU01011698 NCKU01011636 RWS00346.1 RWS00376.1 CP012524 ALC41579.1 CH940648 EDW60118.1 CH916375 EDV98426.1 DQ440303 CH477663 ABF18336.1 EAT37572.1 JXUM01241297 GAPW01006592 KQ635200 JAC07006.1 KXJ62521.1 GL768055 EFZ12794.1 KQ434899 KZC11034.1 GGFM01003726 MBW24477.1 GGFK01010926 MBW44247.1 ADMH02000766 ETN65110.1 CCAG010002640 KK854300 PTY14364.1 GAPW01006721 JAC06877.1 CH933808 EDW09628.1 GFDL01009529 JAV25516.1 DS232085 EDS34396.1 ATLV01027083 KE525423 KFB53856.1 APCN01003547 KT755167 ALS05001.1 GDKW01001976 JAI54619.1 ACPB03008858 GAHY01001255 JAA76255.1 AXCM01001000 GAMD01002973 JAA98617.1 GBBI01001073 JAC17639.1 BT076368 ACO10792.1 GFTR01001512 JAW14914.1 BT077230 ACO11654.1 GECL01001870 JAP04254.1 GBGD01003618 JAC85271.1 HACA01017343 CDW34704.1 UFQS01002647 UFQT01002647 SSX14435.1 SSX33842.1 OUUW01000001 SPP75429.1 CM000071 EAL24941.1 CH479181 EDW31723.1 BT080563 ACO14987.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000007151
UP000053268
UP000218220
+ More
UP000000311 UP000054359 UP000027135 UP000078492 UP000009046 UP000078541 UP000078542 UP000007755 UP000078540 UP000005205 UP000053097 UP000279307 UP000008237 UP000000305 UP000075809 UP000076858 UP000235965 UP000075902 UP000007062 UP000001292 UP000000304 UP000000803 UP000007801 UP000007798 UP000183832 UP000092460 UP000002358 UP000285301 UP000092553 UP000076407 UP000008792 UP000091820 UP000001070 UP000008820 UP000069940 UP000249989 UP000076502 UP000000673 UP000092444 UP000069272 UP000009192 UP000002320 UP000092445 UP000075881 UP000030765 UP000075840 UP000076408 UP000075885 UP000015103 UP000075883 UP000075900 UP000075884 UP000268350 UP000001819 UP000008744 UP000075920
UP000000311 UP000054359 UP000027135 UP000078492 UP000009046 UP000078541 UP000078542 UP000007755 UP000078540 UP000005205 UP000053097 UP000279307 UP000008237 UP000000305 UP000075809 UP000076858 UP000235965 UP000075902 UP000007062 UP000001292 UP000000304 UP000000803 UP000007801 UP000007798 UP000183832 UP000092460 UP000002358 UP000285301 UP000092553 UP000076407 UP000008792 UP000091820 UP000001070 UP000008820 UP000069940 UP000249989 UP000076502 UP000000673 UP000092444 UP000069272 UP000009192 UP000002320 UP000092445 UP000075881 UP000030765 UP000075840 UP000076408 UP000075885 UP000015103 UP000075883 UP000075900 UP000075884 UP000268350 UP000001819 UP000008744 UP000075920
SUPFAM
SSF144122
SSF144122
Gene 3D
ProteinModelPortal
Q2F5K3
A0A3S2LV35
I4DJZ5
A0A194RJV3
A0A212FA50
A0A194QH60
+ More
A0A2A4JV82 A0A2H1VDU2 Q9Y0V4 E2AQP5 A0A087U6B3 A0A067QQ30 A0A2R5LHJ0 R4V508 A0A195D9W6 E0VY47 A0A195F540 A0A151IEZ5 F4W525 A0A195BQ48 A0A158NDJ4 A0A026W2F0 E2B619 A0A023G8F3 E9GAU9 A0A023FR62 A0A0K8R6Y7 A0A1Z5LE75 A0A151XEG9 A0A1E1XV82 A0A0P5ELG7 A0A023FP94 A0A023FVH6 A0A1E1X0C6 A0A131Z5G8 A0A224Z4J2 L7LXC6 A0A131XE60 A0A2J7QM11 A0A0C9QYR7 A0A182U576 U5EZ06 A0A293MEQ1 Q7QE31 A0A1B2AKF1 B4I7U7 B4QGJ5 Q9W2D6 B3MIZ0 T1DHV3 A0A0Q9WYL8 A0A1J1HX77 A0A1B0ARR9 K7IMJ3 A0A3S3NJ03 A0A0M3QV06 A0A182WY03 B4LNP8 A0A1A9WV14 B4JVF8 Q1HQZ1 A0A023EDY9 E9J336 A0A154PHR5 A0A2M3Z7I4 A0A2M4ATZ9 W5JQC8 A0A1B0FD43 A0A2R7W3S0 A0A023EB85 A0A182FVZ1 B4KTP5 A0A1Q3FD67 B0WTF8 A0A1A9ZR57 A0A182JQ62 A0A084WUG2 A0A182I623 A0A0U2TLH7 A0A182YAB3 A0A182PN56 A0A0P4VQ74 R4G4L5 A0A182LZ46 T1E892 A0A023F9G4 C1BP39 A0A224XUT1 C1BRK1 A0A0V0G8G8 A0A069DNN5 A0A0K2U8Z5 A0A182RLV6 A0A336LD03 A0A182NP99 A0A3B0JP97 Q292C1 B4G9X1 C1C134 A0A182WE87
A0A2A4JV82 A0A2H1VDU2 Q9Y0V4 E2AQP5 A0A087U6B3 A0A067QQ30 A0A2R5LHJ0 R4V508 A0A195D9W6 E0VY47 A0A195F540 A0A151IEZ5 F4W525 A0A195BQ48 A0A158NDJ4 A0A026W2F0 E2B619 A0A023G8F3 E9GAU9 A0A023FR62 A0A0K8R6Y7 A0A1Z5LE75 A0A151XEG9 A0A1E1XV82 A0A0P5ELG7 A0A023FP94 A0A023FVH6 A0A1E1X0C6 A0A131Z5G8 A0A224Z4J2 L7LXC6 A0A131XE60 A0A2J7QM11 A0A0C9QYR7 A0A182U576 U5EZ06 A0A293MEQ1 Q7QE31 A0A1B2AKF1 B4I7U7 B4QGJ5 Q9W2D6 B3MIZ0 T1DHV3 A0A0Q9WYL8 A0A1J1HX77 A0A1B0ARR9 K7IMJ3 A0A3S3NJ03 A0A0M3QV06 A0A182WY03 B4LNP8 A0A1A9WV14 B4JVF8 Q1HQZ1 A0A023EDY9 E9J336 A0A154PHR5 A0A2M3Z7I4 A0A2M4ATZ9 W5JQC8 A0A1B0FD43 A0A2R7W3S0 A0A023EB85 A0A182FVZ1 B4KTP5 A0A1Q3FD67 B0WTF8 A0A1A9ZR57 A0A182JQ62 A0A084WUG2 A0A182I623 A0A0U2TLH7 A0A182YAB3 A0A182PN56 A0A0P4VQ74 R4G4L5 A0A182LZ46 T1E892 A0A023F9G4 C1BP39 A0A224XUT1 C1BRK1 A0A0V0G8G8 A0A069DNN5 A0A0K2U8Z5 A0A182RLV6 A0A336LD03 A0A182NP99 A0A3B0JP97 Q292C1 B4G9X1 C1C134 A0A182WE87
PDB
2BSK
E-value=4.61872e-22,
Score=251
Ontologies
GO
PANTHER
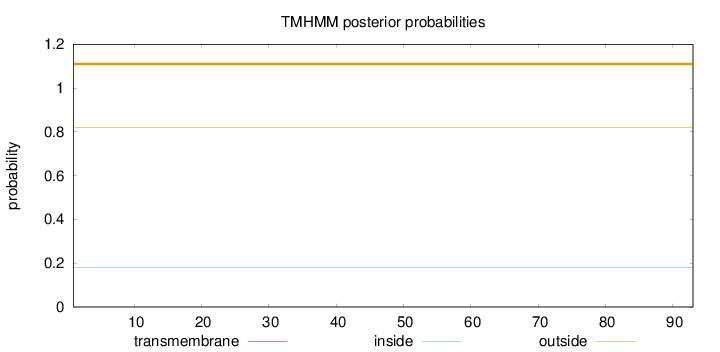
Topology
Subcellular location
Mitochondrion inner membrane
Length:
93
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.18133
outside
1 - 93
Population Genetic Test Statistics
Pi
239.624837
Theta
173.733504
Tajima's D
1.120728
CLR
18.220431
CSRT
0.690115494225289
Interpretation
Uncertain
