Gene
KWMTBOMO14413
Pre Gene Modal
BGIBMGA000121
Annotation
PREDICTED:_GPI_mannosyltransferase_1_[Amyelois_transitella]
Full name
GPI mannosyltransferase 1
Alternative Name
GPI mannosyltransferase I
Phosphatidylinositol-glycan biosynthesis class M protein
Phosphatidylinositol-glycan biosynthesis class M protein
Location in the cell
PlasmaMembrane Reliability : 4.899
Sequence
CDS
ATGGATCGCGTATCGAAATTTATCGATCTATCATTCGGGAACCACATTTGTATCGGTCTGACCGTGAGATTGGTGTTCATATTATACGGGAATATACACGATAGGTTTTCCGTAGTACCATACACTGATGTTGATTACAAAGTTTTCACGGACGCTGCGAGACATTTGGTGGAGGGTAACTCGCCCTATCTCCGGCACACTTACAGATACAGCCCTTTGATCGCGTATCTAATGATACCTAACATTATTGTCGGGAGAGAATTCGGAAAAATCCTTTTTTCCGTTTTCGATATATTTCTGGCGGCTGCCGTCAAATCTCTGGTCGAACGGCAATTGGGCGTCGCGAAATATGCTTCCGTCACAGCTAAATACTGTGCCTTATCTTGGCTCTACAATCCTCTAAGTATAGGCATCTCGACTCGAGGCAATGCCGATTCATTCCCGTGTTTCTTGATCGTCCTATCGATATACTTCCTTCAGTCGAACGTATTTAAAAATTCTATAACCAACATCGTCCTATCAGGCTTCTTTTTAGGTTCCGCAATACATCTACGCTTGTACCCGTTAGCCTTCACTTTCCCTATGTATCTATCTTTAGGAAAATATAAGATAAATCGTAATACCCCAATCAAAGAGGGTCTAGTTTCATTGCTACCCAACAAGAACCAAATGATTTTAGCCTTTAGCTGCGTTACAAGTCTTTTATTTATAACGTACGCTATGTACTTGAGATACGGTTACGAATTCTTATTCGAAACATATATTTATCATCTCCTCAGGAAAGACACTAGGCACAATTTCTCTATACTATTCTACTATACGTATTTAACGAAAGATCAGATGTCATTTGATATTGTCAAAATATTGGCACAGGTTTCCAAGTGCGTTCTACTTTTCCTGCTGAGTCTGAATTACGGTTGTGACCCAAAAACATTACCGTTCGCAATGTTCGCGCAAGCCGTCGTGCTGGTCGCTTATAACAGCGTCATGACATCACAGTACTTTATTTGGTTCCTCTCGTTATTACCATTGGTATTAAAAGACTTCAAAATGGAACCAGGTAAAATTGTTTACTTGGTTTTCCTCTGGATAGCGACTCAATGTGCTTGGTTGTTTTTCGCGTACCAATTAGAGTTCAAATGCAGGCAGGTTTTCATGTTGATCTGGTGTCAGAGCGTTATGTTCTATGGCAGTAATGTTTTTATACTGACGCAGCTTATAAAGTACTACCGCCCCGGCTACGCATTCGGGATAAACGAATCTGGCAAAGGAATAAAAGAGAAATGA
Protein
MDRVSKFIDLSFGNHICIGLTVRLVFILYGNIHDRFSVVPYTDVDYKVFTDAARHLVEGNSPYLRHTYRYSPLIAYLMIPNIIVGREFGKILFSVFDIFLAAAVKSLVERQLGVAKYASVTAKYCALSWLYNPLSIGISTRGNADSFPCFLIVLSIYFLQSNVFKNSITNIVLSGFFLGSAIHLRLYPLAFTFPMYLSLGKYKINRNTPIKEGLVSLLPNKNQMILAFSCVTSLLFITYAMYLRYGYEFLFETYIYHLLRKDTRHNFSILFYYTYLTKDQMSFDIVKILAQVSKCVLLFLLSLNYGCDPKTLPFAMFAQAVVLVAYNSVMTSQYFIWFLSLLPLVLKDFKMEPGKIVYLVFLWIATQCAWLFFAYQLEFKCRQVFMLIWCQSVMFYGSNVFILTQLIKYYRPGYAFGINESGKGIKEK
Summary
Description
Mannosyltransferase involved in glycosylphosphatidylinositol-anchor biosynthesis. Transfers the first alpha-1,4-mannose to GlcN-acyl-PI during GPI precursor assembly.
Mannosyltransferase involved in glycosylphosphatidylinositol-anchor biosynthesis. Transfers the first alpha-1,4-mannose to GlcN-acyl-PI during GPI precursor assembly (By similarity).
Mannosyltransferase involved in glycosylphosphatidylinositol-anchor biosynthesis. Transfers the first alpha-1,4-mannose to GlcN-acyl-PI during GPI precursor assembly (By similarity).
Similarity
Belongs to the PIGM family.
Keywords
Complete proteome
Endoplasmic reticulum
Glycosyltransferase
GPI-anchor biosynthesis
Membrane
Reference proteome
Transferase
Transmembrane
Transmembrane helix
Feature
chain GPI mannosyltransferase 1
Uniprot
H9IS94
A0A1E1W669
A0A194QH28
A0A0L7LJ24
A0A212FA37
A0A026W2R9
+ More
A0A158NC27 A0A2J7R9A6 F4X8B0 A0A067QTY4 A0A1B6JC24 A0A1B6HQE1 A0A151IMB5 A0A336N1M2 A0A146LGX8 A0A336N8Y0 A0A1L8E1N4 A0A1L8E1A0 A0A069DT57 A0A1B6JC83 A0A1B6J9R3 A0A1B6JUU9 A0A1B6GNV9 E2C426 A0A224XHY9 A0A1W4WVN6 K7IWX6 A0A0M3QVF1 B4MIV5 A0A023F7R0 A0A1A9WV10 A0A1A9ZQM3 B4JVN4 A0A1I8Q911 A0A2P8Y7V2 A0A1B6DAR5 E0VBV0 J9JT12 W8BE37 A0A2S2PYY3 A0A034V763 A0A034V3U1 A0A2P2HXA1 A0A0K8WBT9 B4KML5 B4MCL3 A0A2R7X2G4 T1HNQ1 A0A0P4W4B8 R4G564 A0A2S2NUM9 A0A131Y4G8 B7PCK7 A0A0P4W714 A0A0H5R619 V5HJR5 A0A3M6UWS7 B3RIQ9 A0A1I8MJC0 A0A0A1XT11 A7STD8 R7U587 A0A369SL93 W4YY66 R7UFS0 A0A1Y1HII1 V4A763 F4QEX9 A0A3S1I0V8 A0A3B4TM41 G3NB04 A0A2I4CZY0 A0A3B4WDI1 A0A1A9X9R9 A0A1B0C0J8 Q54IA4 A0A060WQB7 A0A2U9CY02 E6ZI90 C1C161 A0A3Q3MV71 A0A0G4J309 A0A3P9NUS1 A0A3Q1EZM5 A0A0N8JVH2 A0A146NT24 I3KNE3 A0A3P8SG34 A0A3B3CH26 A0A0P5R3E3 A0A0N8EED7 H2LKX9 A0A3Q2WKU1 A0A0P5WQV3 A0A3Q0RS24 A0A0P5XIR5 A0A3P9DH51 H3CTL4 A0A3Q2PS46
A0A158NC27 A0A2J7R9A6 F4X8B0 A0A067QTY4 A0A1B6JC24 A0A1B6HQE1 A0A151IMB5 A0A336N1M2 A0A146LGX8 A0A336N8Y0 A0A1L8E1N4 A0A1L8E1A0 A0A069DT57 A0A1B6JC83 A0A1B6J9R3 A0A1B6JUU9 A0A1B6GNV9 E2C426 A0A224XHY9 A0A1W4WVN6 K7IWX6 A0A0M3QVF1 B4MIV5 A0A023F7R0 A0A1A9WV10 A0A1A9ZQM3 B4JVN4 A0A1I8Q911 A0A2P8Y7V2 A0A1B6DAR5 E0VBV0 J9JT12 W8BE37 A0A2S2PYY3 A0A034V763 A0A034V3U1 A0A2P2HXA1 A0A0K8WBT9 B4KML5 B4MCL3 A0A2R7X2G4 T1HNQ1 A0A0P4W4B8 R4G564 A0A2S2NUM9 A0A131Y4G8 B7PCK7 A0A0P4W714 A0A0H5R619 V5HJR5 A0A3M6UWS7 B3RIQ9 A0A1I8MJC0 A0A0A1XT11 A7STD8 R7U587 A0A369SL93 W4YY66 R7UFS0 A0A1Y1HII1 V4A763 F4QEX9 A0A3S1I0V8 A0A3B4TM41 G3NB04 A0A2I4CZY0 A0A3B4WDI1 A0A1A9X9R9 A0A1B0C0J8 Q54IA4 A0A060WQB7 A0A2U9CY02 E6ZI90 C1C161 A0A3Q3MV71 A0A0G4J309 A0A3P9NUS1 A0A3Q1EZM5 A0A0N8JVH2 A0A146NT24 I3KNE3 A0A3P8SG34 A0A3B3CH26 A0A0P5R3E3 A0A0N8EED7 H2LKX9 A0A3Q2WKU1 A0A0P5WQV3 A0A3Q0RS24 A0A0P5XIR5 A0A3P9DH51 H3CTL4 A0A3Q2PS46
EC Number
2.4.1.-
Pubmed
19121390
26354079
26227816
22118469
24508170
30249741
+ More
21347285 21719571 24845553 26823975 26334808 20798317 20075255 17994087 25474469 29403074 20566863 24495485 25348373 27129103 25765539 30382153 18719581 25315136 25830018 17615350 23254933 30042472 24865297 21757610 15875012 24755649 25186727 29451363 17554307 15496914
21347285 21719571 24845553 26823975 26334808 20798317 20075255 17994087 25474469 29403074 20566863 24495485 25348373 27129103 25765539 30382153 18719581 25315136 25830018 17615350 23254933 30042472 24865297 21757610 15875012 24755649 25186727 29451363 17554307 15496914
EMBL
BABH01024106
GDQN01008572
JAT82482.1
KQ458859
KPJ04848.1
JTDY01000885
+ More
KOB75558.1 AGBW02009528 OWR50588.1 KK107459 QOIP01000004 EZA50380.1 RLU23715.1 ADTU01011316 NEVH01006600 PNF37418.1 GL888932 EGI57180.1 KK852954 KDR13289.1 GECU01010876 JAS96830.1 GECU01030814 JAS76892.1 KQ977059 KYN06013.1 UFQS01005052 UFQT01005052 SSX16682.1 SSX35875.1 GDHC01012417 JAQ06212.1 UFQS01003696 UFQT01003696 SSX15864.1 SSX35208.1 GFDF01001602 JAV12482.1 GFDF01001603 JAV12481.1 GBGD01001754 JAC87135.1 GECU01010907 GECU01002551 JAS96799.1 JAT05156.1 GECU01011837 JAS95869.1 GECU01036594 GECU01028258 GECU01004726 JAS71112.1 JAS79448.1 JAT02981.1 GECZ01005655 JAS64114.1 GL452364 EFN77412.1 GFTR01005762 JAW10664.1 AAZX01009385 CP012524 ALC42309.1 CH963719 EDW72044.2 GBBI01001444 JAC17268.1 CH916375 EDV98502.1 PYGN01000823 PSN40336.1 GEDC01014566 JAS22732.1 DS235042 EEB10856.1 ABLF02009097 GAMC01011287 JAB95268.1 GGMS01001533 MBY70736.1 GAKP01020993 GAKP01020990 JAC37962.1 GAKP01020992 GAKP01020991 JAC37961.1 IACF01000655 LAB66417.1 GDHF01003688 GDHF01000021 JAI48626.1 JAI52293.1 CH933808 EDW10862.1 CH940659 EDW71401.1 KK856473 PTY25851.1 ACPB03007520 GDKW01000408 JAI56187.1 GAHY01000181 JAA77329.1 GGMR01007817 MBY20436.1 GEFM01002850 JAP72946.1 ABJB010474095 DS684369 EEC04329.1 GDRN01086856 JAI61114.1 HACM01008839 CRZ09281.1 GANP01010725 JAB73743.1 RCHS01000548 RMX58070.1 DS985241 EDV29023.1 GBXI01000106 JAD14186.1 DS469794 EDO33030.1 AMQN01026870 KB307513 ELT98841.1 NOWV01000004 RDD47244.1 AAGJ04003702 AAGJ04003703 AMQN01001554 KB303857 ELU02643.1 DF236959 GAQ78300.1 KB202325 ESO90830.1 GL883029 EGG14186.1 RQTK01000042 RUS90035.1 JXJN01023653 AAFI02000126 FR904577 CDQ67229.1 CP026263 AWP20910.1 FQ310508 CBN81774.1 BT080590 ACO15014.1 CDSF01000122 OVEO01000010 CEP01960.1 SPQ98819.1 JARO02014602 KPP58129.1 GCES01151444 JAQ34878.1 AERX01067164 GDIQ01126041 JAL25685.1 GDIQ01034972 LRGB01000190 JAN59765.1 KZS20491.1 GDIP01082906 JAM20809.1 GDIP01071563 JAM32152.1
KOB75558.1 AGBW02009528 OWR50588.1 KK107459 QOIP01000004 EZA50380.1 RLU23715.1 ADTU01011316 NEVH01006600 PNF37418.1 GL888932 EGI57180.1 KK852954 KDR13289.1 GECU01010876 JAS96830.1 GECU01030814 JAS76892.1 KQ977059 KYN06013.1 UFQS01005052 UFQT01005052 SSX16682.1 SSX35875.1 GDHC01012417 JAQ06212.1 UFQS01003696 UFQT01003696 SSX15864.1 SSX35208.1 GFDF01001602 JAV12482.1 GFDF01001603 JAV12481.1 GBGD01001754 JAC87135.1 GECU01010907 GECU01002551 JAS96799.1 JAT05156.1 GECU01011837 JAS95869.1 GECU01036594 GECU01028258 GECU01004726 JAS71112.1 JAS79448.1 JAT02981.1 GECZ01005655 JAS64114.1 GL452364 EFN77412.1 GFTR01005762 JAW10664.1 AAZX01009385 CP012524 ALC42309.1 CH963719 EDW72044.2 GBBI01001444 JAC17268.1 CH916375 EDV98502.1 PYGN01000823 PSN40336.1 GEDC01014566 JAS22732.1 DS235042 EEB10856.1 ABLF02009097 GAMC01011287 JAB95268.1 GGMS01001533 MBY70736.1 GAKP01020993 GAKP01020990 JAC37962.1 GAKP01020992 GAKP01020991 JAC37961.1 IACF01000655 LAB66417.1 GDHF01003688 GDHF01000021 JAI48626.1 JAI52293.1 CH933808 EDW10862.1 CH940659 EDW71401.1 KK856473 PTY25851.1 ACPB03007520 GDKW01000408 JAI56187.1 GAHY01000181 JAA77329.1 GGMR01007817 MBY20436.1 GEFM01002850 JAP72946.1 ABJB010474095 DS684369 EEC04329.1 GDRN01086856 JAI61114.1 HACM01008839 CRZ09281.1 GANP01010725 JAB73743.1 RCHS01000548 RMX58070.1 DS985241 EDV29023.1 GBXI01000106 JAD14186.1 DS469794 EDO33030.1 AMQN01026870 KB307513 ELT98841.1 NOWV01000004 RDD47244.1 AAGJ04003702 AAGJ04003703 AMQN01001554 KB303857 ELU02643.1 DF236959 GAQ78300.1 KB202325 ESO90830.1 GL883029 EGG14186.1 RQTK01000042 RUS90035.1 JXJN01023653 AAFI02000126 FR904577 CDQ67229.1 CP026263 AWP20910.1 FQ310508 CBN81774.1 BT080590 ACO15014.1 CDSF01000122 OVEO01000010 CEP01960.1 SPQ98819.1 JARO02014602 KPP58129.1 GCES01151444 JAQ34878.1 AERX01067164 GDIQ01126041 JAL25685.1 GDIQ01034972 LRGB01000190 JAN59765.1 KZS20491.1 GDIP01082906 JAM20809.1 GDIP01071563 JAM32152.1
Proteomes
UP000005204
UP000053268
UP000037510
UP000007151
UP000053097
UP000279307
+ More
UP000005205 UP000235965 UP000007755 UP000027135 UP000078542 UP000008237 UP000192223 UP000002358 UP000092553 UP000007798 UP000091820 UP000092445 UP000001070 UP000095300 UP000245037 UP000009046 UP000007819 UP000009192 UP000008792 UP000015103 UP000001555 UP000275408 UP000009022 UP000095301 UP000001593 UP000014760 UP000253843 UP000007110 UP000054558 UP000030746 UP000007797 UP000271974 UP000261420 UP000007635 UP000192220 UP000261360 UP000092443 UP000092460 UP000002195 UP000193380 UP000246464 UP000261640 UP000039324 UP000290189 UP000242638 UP000257200 UP000034805 UP000192224 UP000005207 UP000265080 UP000261560 UP000076858 UP000001038 UP000264840 UP000261340 UP000265160 UP000007303 UP000265000
UP000005205 UP000235965 UP000007755 UP000027135 UP000078542 UP000008237 UP000192223 UP000002358 UP000092553 UP000007798 UP000091820 UP000092445 UP000001070 UP000095300 UP000245037 UP000009046 UP000007819 UP000009192 UP000008792 UP000015103 UP000001555 UP000275408 UP000009022 UP000095301 UP000001593 UP000014760 UP000253843 UP000007110 UP000054558 UP000030746 UP000007797 UP000271974 UP000261420 UP000007635 UP000192220 UP000261360 UP000092443 UP000092460 UP000002195 UP000193380 UP000246464 UP000261640 UP000039324 UP000290189 UP000242638 UP000257200 UP000034805 UP000192224 UP000005207 UP000265080 UP000261560 UP000076858 UP000001038 UP000264840 UP000261340 UP000265160 UP000007303 UP000265000
Pfam
PF05007 Mannosyl_trans
Interpro
IPR007704
PIG-M
ProteinModelPortal
H9IS94
A0A1E1W669
A0A194QH28
A0A0L7LJ24
A0A212FA37
A0A026W2R9
+ More
A0A158NC27 A0A2J7R9A6 F4X8B0 A0A067QTY4 A0A1B6JC24 A0A1B6HQE1 A0A151IMB5 A0A336N1M2 A0A146LGX8 A0A336N8Y0 A0A1L8E1N4 A0A1L8E1A0 A0A069DT57 A0A1B6JC83 A0A1B6J9R3 A0A1B6JUU9 A0A1B6GNV9 E2C426 A0A224XHY9 A0A1W4WVN6 K7IWX6 A0A0M3QVF1 B4MIV5 A0A023F7R0 A0A1A9WV10 A0A1A9ZQM3 B4JVN4 A0A1I8Q911 A0A2P8Y7V2 A0A1B6DAR5 E0VBV0 J9JT12 W8BE37 A0A2S2PYY3 A0A034V763 A0A034V3U1 A0A2P2HXA1 A0A0K8WBT9 B4KML5 B4MCL3 A0A2R7X2G4 T1HNQ1 A0A0P4W4B8 R4G564 A0A2S2NUM9 A0A131Y4G8 B7PCK7 A0A0P4W714 A0A0H5R619 V5HJR5 A0A3M6UWS7 B3RIQ9 A0A1I8MJC0 A0A0A1XT11 A7STD8 R7U587 A0A369SL93 W4YY66 R7UFS0 A0A1Y1HII1 V4A763 F4QEX9 A0A3S1I0V8 A0A3B4TM41 G3NB04 A0A2I4CZY0 A0A3B4WDI1 A0A1A9X9R9 A0A1B0C0J8 Q54IA4 A0A060WQB7 A0A2U9CY02 E6ZI90 C1C161 A0A3Q3MV71 A0A0G4J309 A0A3P9NUS1 A0A3Q1EZM5 A0A0N8JVH2 A0A146NT24 I3KNE3 A0A3P8SG34 A0A3B3CH26 A0A0P5R3E3 A0A0N8EED7 H2LKX9 A0A3Q2WKU1 A0A0P5WQV3 A0A3Q0RS24 A0A0P5XIR5 A0A3P9DH51 H3CTL4 A0A3Q2PS46
A0A158NC27 A0A2J7R9A6 F4X8B0 A0A067QTY4 A0A1B6JC24 A0A1B6HQE1 A0A151IMB5 A0A336N1M2 A0A146LGX8 A0A336N8Y0 A0A1L8E1N4 A0A1L8E1A0 A0A069DT57 A0A1B6JC83 A0A1B6J9R3 A0A1B6JUU9 A0A1B6GNV9 E2C426 A0A224XHY9 A0A1W4WVN6 K7IWX6 A0A0M3QVF1 B4MIV5 A0A023F7R0 A0A1A9WV10 A0A1A9ZQM3 B4JVN4 A0A1I8Q911 A0A2P8Y7V2 A0A1B6DAR5 E0VBV0 J9JT12 W8BE37 A0A2S2PYY3 A0A034V763 A0A034V3U1 A0A2P2HXA1 A0A0K8WBT9 B4KML5 B4MCL3 A0A2R7X2G4 T1HNQ1 A0A0P4W4B8 R4G564 A0A2S2NUM9 A0A131Y4G8 B7PCK7 A0A0P4W714 A0A0H5R619 V5HJR5 A0A3M6UWS7 B3RIQ9 A0A1I8MJC0 A0A0A1XT11 A7STD8 R7U587 A0A369SL93 W4YY66 R7UFS0 A0A1Y1HII1 V4A763 F4QEX9 A0A3S1I0V8 A0A3B4TM41 G3NB04 A0A2I4CZY0 A0A3B4WDI1 A0A1A9X9R9 A0A1B0C0J8 Q54IA4 A0A060WQB7 A0A2U9CY02 E6ZI90 C1C161 A0A3Q3MV71 A0A0G4J309 A0A3P9NUS1 A0A3Q1EZM5 A0A0N8JVH2 A0A146NT24 I3KNE3 A0A3P8SG34 A0A3B3CH26 A0A0P5R3E3 A0A0N8EED7 H2LKX9 A0A3Q2WKU1 A0A0P5WQV3 A0A3Q0RS24 A0A0P5XIR5 A0A3P9DH51 H3CTL4 A0A3Q2PS46
Ontologies
PATHWAY
GO
PANTHER
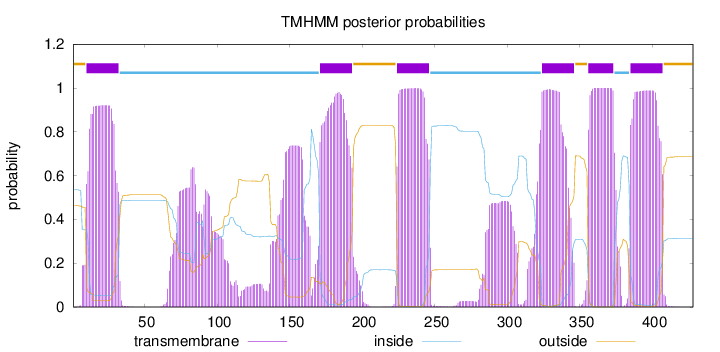
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
428
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
174.7631
Exp number, first 60 AAs:
19.84649
Total prob of N-in:
0.53580
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 170
TMhelix
171 - 193
outside
194 - 223
TMhelix
224 - 246
inside
247 - 323
TMhelix
324 - 346
outside
347 - 355
TMhelix
356 - 373
inside
374 - 384
TMhelix
385 - 407
outside
408 - 428
Population Genetic Test Statistics
Pi
272.089521
Theta
192.602978
Tajima's D
1.218436
CLR
0.000011
CSRT
0.715714214289285
Interpretation
Uncertain
