Gene
KWMTBOMO14412 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000120
Annotation
PREDICTED:_vacuolar_protein_sorting-associated_protein_26B-like_[Amyelois_transitella]
Full name
Vacuolar protein sorting-associated protein 26
Alternative Name
VPS26 protein homolog
Location in the cell
Cytoplasmic Reliability : 2.174 Nuclear Reliability : 2.409
Sequence
CDS
ATGAGCTTTTTCGGGTTCGGTCAGACGGCGGATATCGAAATAGTTTTTGATGATGCTGACAAACGGAAGGTGGCTGAAGTGAAAACGGACGATGGTAAAAAAGAAAAGTTATTGTTGTATTATGACGGTGAAACGGTCTCCGGGAAAGTGAATGTGACGTTGCGGAAACCGGGATCAAAACTTGAACATCAAGGCATCAAGGTCGAACTCATAGGTCAGATTGAATTATTCTACGACAGAGGTAATCATCACGAATTCATATCTCTGGTTAAAGAACTAGCGCGTCCGGGCGACCTGCTTCAACACACGTCGTATCCTTTCGAGTTTGCTAATGTCGAAAAACCATACGAAGTATATACAGGCTCTAATGTCAGGTTGCGGTATTTCCTGCGGGCGACAATCGTCCGGCGCCTCACTGACATCACCAAGGAGGTGGATATTGCCGTGCACACGCTCTGTAGTTATCCGGACGTCCTTAATTCTATAAAAATGGAAGTAGGCATAGAAGACTGTCTGCACATAGAGTTTGAATATAACAAATCAAAATATCATTTGAAAGACGTTATTGTTGGTAAAATTTATTTCTTACTGGTACGTATAAAAATCAAGCACATGGAAATATCAATAATCAAGAGAGAAACCACTGGATCTGGTCCCAATACGTTTACAGAAAATGAAACTGTAGCAAAGTATGAGATAATGGATGGTGCGCCAGTCAGGGGTGAAAGTATTCCTATCAGAGTGTTTTTGGCTGGCTATGACCTGACTCCGACTATGAGGGACATTAACAACAAATTTTCTGTCAGATACTACCTTAATTTAGTTTTGATGGACACTGAAGATCGTCGCTATTTCAAACAACAAGAGGTGATATTATGGAGGAAGAGTGACAAGTCCAGATTACCCTTGCATCCTCATCACCCACAAACAGTGTCATATCAAGGCCATCAGTCATTAAGAAAACAAAGTGTTTCCAGTGATGATAATTCAGCTCGAGCAACACCATCAAATCCTGATCCTGAGAATGTAATGCAGAGATCAGTGTCCCCCTCAATGCCTGAAAAACAGAATGGACCCTTACAAATGGAGAGAGAAAAACCTGAAGCATTCATAGACAAATTAGCAGGTGCTCATATTAATGAAAATGATACTAACGATACGAGTGATGAGATTGAAGAACCGAAACCCGTCGAAAAACTTCCCGTAGTGGATAAACCTTTAATCGCCGAAAAACCTCAGATTGCTGAAAAACCTACCCTAAACCGACCTGAGCCTGCAGGGAGTCCGAATCAATAG
Protein
MSFFGFGQTADIEIVFDDADKRKVAEVKTDDGKKEKLLLYYDGETVSGKVNVTLRKPGSKLEHQGIKVELIGQIELFYDRGNHHEFISLVKELARPGDLLQHTSYPFEFANVEKPYEVYTGSNVRLRYFLRATIVRRLTDITKEVDIAVHTLCSYPDVLNSIKMEVGIEDCLHIEFEYNKSKYHLKDVIVGKIYFLLVRIKIKHMEISIIKRETTGSGPNTFTENETVAKYEIMDGAPVRGESIPIRVFLAGYDLTPTMRDINNKFSVRYYLNLVLMDTEDRRYFKQQEVILWRKSDKSRLPLHPHHPQTVSYQGHQSLRKQSVSSDDNSARATPSNPDPENVMQRSVSPSMPEKQNGPLQMEREKPEAFIDKLAGAHINENDTNDTSDEIEEPKPVEKLPVVDKPLIAEKPQIAEKPTLNRPEPAGSPNQ
Summary
Description
Component of the retromer complex which acts in conjunction with wingless (wg) and clathrin-mediated endocytosis to sustain a wntless (wls) traffic loop. This loop encompasses the Golgi, the cell surface, an endocytic compartment and a retrograde route leading back to the Golgi, thereby enabling wls to direct wg secretion. The hh and dpp signaling pathways do not require the retromer complex suggesting that it does not play a general role in exocytosis.
Subunit
Component of the retromer complex, composed of Vps26 and Vps35.
Similarity
Belongs to the VPS26 family.
Keywords
Complete proteome
Cytoplasm
Membrane
Protein transport
Reference proteome
Transport
Wnt signaling pathway
Feature
chain Vacuolar protein sorting-associated protein 26
Uniprot
H9IS93
A0A2H1WUD2
A0A2A4JV15
A0A0L7LJC9
A0A3S2TG18
A0A194QMZ1
+ More
A0A194RJK5 A0A212FA24 A0A1L8DVC7 A0A336M094 A0A336MUK6 A0A0K8TSB1 U5EWK8 D2A583 A0A0T6BD35 A0A1I8MNP2 A0A2M4AFJ4 A0A1Q3F910 A0A1Q3F8S6 A0A2M3Z479 A0A2M4AFT7 A0A2M4BHM7 A0A2M4BHJ9 A0A2M4BHS7 W5J8V2 D3TNJ9 A0A0L0C435 A0A2M3Z426 A0A1S4G1K5 A0A182XP88 A0A067QYJ0 Q7Q5Y3 A0A2J7Q1J4 A0A0A1WRR9 A0A1A9WA17 A0A182Y2R6 A0A182KNZ4 A0A1A9XM10 A0A034WM28 A0A182I1D8 A0A1Y9HDZ0 A0A182UPZ1 A0A182FEI9 A0A182GF45 A0A1A9Z3Y4 A0A2M3Z558 W8BR63 A0A1W4X737 A0A226CXH6 A0A182JPN5 A0A182LXX7 A0A1A9VWW5 A0A1E1XTV5 A0A182IL17 A0A1J1J444 A0A084WEQ8 A0A182PDS1 A0A023GLK5 G3MM37 A0A1E1X9Z7 A0A0K8W0J3 A0A0M4FAP9 A0A023FIT6 Q16KJ7 A0A1B6L663 A0A1B6FGW5 B7PQ07 V5H0U0 V9ILB7 A0A088A5E3 A0A1B6H6C5 A0A1S3HY72 A0A2A3E6Q0 A0A0M9A5I2 A0A0C9RVK4 A0A3L8DE08 A0A0V0GA44 T1HIG2 A0A069DYT0 A0A023FWI7 A0A2R5LJQ5 A0A224XEF2 B4PY70 V3ZKD4 A0A151I0X6 Q9W552 B4I9J7 A0A0L7R868
A0A194RJK5 A0A212FA24 A0A1L8DVC7 A0A336M094 A0A336MUK6 A0A0K8TSB1 U5EWK8 D2A583 A0A0T6BD35 A0A1I8MNP2 A0A2M4AFJ4 A0A1Q3F910 A0A1Q3F8S6 A0A2M3Z479 A0A2M4AFT7 A0A2M4BHM7 A0A2M4BHJ9 A0A2M4BHS7 W5J8V2 D3TNJ9 A0A0L0C435 A0A2M3Z426 A0A1S4G1K5 A0A182XP88 A0A067QYJ0 Q7Q5Y3 A0A2J7Q1J4 A0A0A1WRR9 A0A1A9WA17 A0A182Y2R6 A0A182KNZ4 A0A1A9XM10 A0A034WM28 A0A182I1D8 A0A1Y9HDZ0 A0A182UPZ1 A0A182FEI9 A0A182GF45 A0A1A9Z3Y4 A0A2M3Z558 W8BR63 A0A1W4X737 A0A226CXH6 A0A182JPN5 A0A182LXX7 A0A1A9VWW5 A0A1E1XTV5 A0A182IL17 A0A1J1J444 A0A084WEQ8 A0A182PDS1 A0A023GLK5 G3MM37 A0A1E1X9Z7 A0A0K8W0J3 A0A0M4FAP9 A0A023FIT6 Q16KJ7 A0A1B6L663 A0A1B6FGW5 B7PQ07 V5H0U0 V9ILB7 A0A088A5E3 A0A1B6H6C5 A0A1S3HY72 A0A2A3E6Q0 A0A0M9A5I2 A0A0C9RVK4 A0A3L8DE08 A0A0V0GA44 T1HIG2 A0A069DYT0 A0A023FWI7 A0A2R5LJQ5 A0A224XEF2 B4PY70 V3ZKD4 A0A151I0X6 Q9W552 B4I9J7 A0A0L7R868
Pubmed
19121390
26227816
26354079
22118469
26369729
18362917
+ More
19820115 25315136 20920257 23761445 20353571 26108605 17510324 24845553 12364791 14747013 17210077 25830018 25244985 20966253 25348373 26483478 24495485 29209593 24438588 22216098 28503490 25765539 30249741 26334808 17994087 17550304 23254933 10731132 12537572 10731137 12537569 18193032
19820115 25315136 20920257 23761445 20353571 26108605 17510324 24845553 12364791 14747013 17210077 25830018 25244985 20966253 25348373 26483478 24495485 29209593 24438588 22216098 28503490 25765539 30249741 26334808 17994087 17550304 23254933 10731132 12537572 10731137 12537569 18193032
EMBL
BABH01024106
ODYU01010731
SOQ56024.1
NWSH01000581
PCG75528.1
JTDY01000885
+ More
KOB75557.1 RSAL01000179 RVE44970.1 KQ458859 KPJ04851.1 KQ460124 KPJ17619.1 AGBW02009528 OWR50587.1 GFDF01003704 JAV10380.1 UFQT01000242 SSX22263.1 UFQS01002031 UFQT01002031 SSX13023.1 SSX32463.1 GDAI01000359 JAI17244.1 GANO01002912 JAB56959.1 KQ971345 EFA05114.1 LJIG01001689 KRT85251.1 GGFK01006233 MBW39554.1 GFDL01010995 JAV24050.1 GFDL01011074 JAV23971.1 GGFM01002552 MBW23303.1 GGFK01006261 MBW39582.1 GGFJ01003401 MBW52542.1 GGFJ01003371 MBW52512.1 GGFJ01003370 MBW52511.1 ADMH02002097 ETN59229.1 CCAG010023252 EZ423001 ADD19277.1 JRES01000940 KNC27001.1 GGFM01002515 MBW23266.1 KK852822 KDR15525.1 AAAB01008960 EAA10832.4 EDO63799.1 NEVH01019406 PNF22447.1 GBXI01013077 JAD01215.1 GAKP01002326 JAC56626.1 APCN01000054 JXUM01059195 KQ562039 KXJ76842.1 GGFM01002908 MBW23659.1 GAMC01002765 JAC03791.1 LNIX01000061 OXA37238.1 AXCM01003909 GFAA01000706 JAU02729.1 CVRI01000070 CRL07175.1 ATLV01023245 KE525341 KFB48702.1 GBBM01001718 JAC33700.1 JO842938 AEO34555.1 GFAC01003146 JAT96042.1 GDHF01007710 JAI44604.1 CP012528 ALC49520.1 GBBK01002975 JAC21507.1 CH477953 EAT34835.1 GEBQ01020971 GEBQ01020769 JAT19006.1 JAT19208.1 GECZ01030060 GECZ01020342 GECZ01008133 JAS39709.1 JAS49427.1 JAS61636.1 ABJB010160378 ABJB010289233 ABJB010500975 DS762323 EEC08679.1 GANP01007873 GEFM01003773 JAB76595.1 JAP72023.1 JR053141 AEY61860.1 GECU01037460 GECU01033474 GECU01032347 GECU01018781 GECU01016243 GECU01013012 GECU01000961 JAS70246.1 JAS74232.1 JAS75359.1 JAS88925.1 JAS91463.1 JAS94694.1 JAT06746.1 KZ288364 PBC26896.1 KQ435729 KOX77647.1 GBYB01012975 JAG82742.1 QOIP01000009 RLU18634.1 GECL01001858 JAP04266.1 ACPB03010282 GBGD01001855 JAC87034.1 GBBL01001456 JAC25864.1 GGLE01005610 MBY09736.1 GFTR01005570 JAW10856.1 CM000162 EDX00943.1 KB202199 ESO91783.1 KQ976600 KYM79464.1 AE014298 AL021106 BT023502 AY060842 CH480825 EDW43878.1 KQ414633 KOC67075.1
KOB75557.1 RSAL01000179 RVE44970.1 KQ458859 KPJ04851.1 KQ460124 KPJ17619.1 AGBW02009528 OWR50587.1 GFDF01003704 JAV10380.1 UFQT01000242 SSX22263.1 UFQS01002031 UFQT01002031 SSX13023.1 SSX32463.1 GDAI01000359 JAI17244.1 GANO01002912 JAB56959.1 KQ971345 EFA05114.1 LJIG01001689 KRT85251.1 GGFK01006233 MBW39554.1 GFDL01010995 JAV24050.1 GFDL01011074 JAV23971.1 GGFM01002552 MBW23303.1 GGFK01006261 MBW39582.1 GGFJ01003401 MBW52542.1 GGFJ01003371 MBW52512.1 GGFJ01003370 MBW52511.1 ADMH02002097 ETN59229.1 CCAG010023252 EZ423001 ADD19277.1 JRES01000940 KNC27001.1 GGFM01002515 MBW23266.1 KK852822 KDR15525.1 AAAB01008960 EAA10832.4 EDO63799.1 NEVH01019406 PNF22447.1 GBXI01013077 JAD01215.1 GAKP01002326 JAC56626.1 APCN01000054 JXUM01059195 KQ562039 KXJ76842.1 GGFM01002908 MBW23659.1 GAMC01002765 JAC03791.1 LNIX01000061 OXA37238.1 AXCM01003909 GFAA01000706 JAU02729.1 CVRI01000070 CRL07175.1 ATLV01023245 KE525341 KFB48702.1 GBBM01001718 JAC33700.1 JO842938 AEO34555.1 GFAC01003146 JAT96042.1 GDHF01007710 JAI44604.1 CP012528 ALC49520.1 GBBK01002975 JAC21507.1 CH477953 EAT34835.1 GEBQ01020971 GEBQ01020769 JAT19006.1 JAT19208.1 GECZ01030060 GECZ01020342 GECZ01008133 JAS39709.1 JAS49427.1 JAS61636.1 ABJB010160378 ABJB010289233 ABJB010500975 DS762323 EEC08679.1 GANP01007873 GEFM01003773 JAB76595.1 JAP72023.1 JR053141 AEY61860.1 GECU01037460 GECU01033474 GECU01032347 GECU01018781 GECU01016243 GECU01013012 GECU01000961 JAS70246.1 JAS74232.1 JAS75359.1 JAS88925.1 JAS91463.1 JAS94694.1 JAT06746.1 KZ288364 PBC26896.1 KQ435729 KOX77647.1 GBYB01012975 JAG82742.1 QOIP01000009 RLU18634.1 GECL01001858 JAP04266.1 ACPB03010282 GBGD01001855 JAC87034.1 GBBL01001456 JAC25864.1 GGLE01005610 MBY09736.1 GFTR01005570 JAW10856.1 CM000162 EDX00943.1 KB202199 ESO91783.1 KQ976600 KYM79464.1 AE014298 AL021106 BT023502 AY060842 CH480825 EDW43878.1 KQ414633 KOC67075.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000053268
UP000053240
+ More
UP000007151 UP000007266 UP000095301 UP000000673 UP000092444 UP000037069 UP000076407 UP000027135 UP000007062 UP000235965 UP000091820 UP000076408 UP000075882 UP000092443 UP000075840 UP000075900 UP000075903 UP000069272 UP000069940 UP000249989 UP000092445 UP000192223 UP000198287 UP000075881 UP000075883 UP000078200 UP000075880 UP000183832 UP000030765 UP000075885 UP000092553 UP000008820 UP000001555 UP000005203 UP000085678 UP000242457 UP000053105 UP000279307 UP000015103 UP000002282 UP000030746 UP000078540 UP000000803 UP000001292 UP000053825
UP000007151 UP000007266 UP000095301 UP000000673 UP000092444 UP000037069 UP000076407 UP000027135 UP000007062 UP000235965 UP000091820 UP000076408 UP000075882 UP000092443 UP000075840 UP000075900 UP000075903 UP000069272 UP000069940 UP000249989 UP000092445 UP000192223 UP000198287 UP000075881 UP000075883 UP000078200 UP000075880 UP000183832 UP000030765 UP000075885 UP000092553 UP000008820 UP000001555 UP000005203 UP000085678 UP000242457 UP000053105 UP000279307 UP000015103 UP000002282 UP000030746 UP000078540 UP000000803 UP000001292 UP000053825
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
H9IS93
A0A2H1WUD2
A0A2A4JV15
A0A0L7LJC9
A0A3S2TG18
A0A194QMZ1
+ More
A0A194RJK5 A0A212FA24 A0A1L8DVC7 A0A336M094 A0A336MUK6 A0A0K8TSB1 U5EWK8 D2A583 A0A0T6BD35 A0A1I8MNP2 A0A2M4AFJ4 A0A1Q3F910 A0A1Q3F8S6 A0A2M3Z479 A0A2M4AFT7 A0A2M4BHM7 A0A2M4BHJ9 A0A2M4BHS7 W5J8V2 D3TNJ9 A0A0L0C435 A0A2M3Z426 A0A1S4G1K5 A0A182XP88 A0A067QYJ0 Q7Q5Y3 A0A2J7Q1J4 A0A0A1WRR9 A0A1A9WA17 A0A182Y2R6 A0A182KNZ4 A0A1A9XM10 A0A034WM28 A0A182I1D8 A0A1Y9HDZ0 A0A182UPZ1 A0A182FEI9 A0A182GF45 A0A1A9Z3Y4 A0A2M3Z558 W8BR63 A0A1W4X737 A0A226CXH6 A0A182JPN5 A0A182LXX7 A0A1A9VWW5 A0A1E1XTV5 A0A182IL17 A0A1J1J444 A0A084WEQ8 A0A182PDS1 A0A023GLK5 G3MM37 A0A1E1X9Z7 A0A0K8W0J3 A0A0M4FAP9 A0A023FIT6 Q16KJ7 A0A1B6L663 A0A1B6FGW5 B7PQ07 V5H0U0 V9ILB7 A0A088A5E3 A0A1B6H6C5 A0A1S3HY72 A0A2A3E6Q0 A0A0M9A5I2 A0A0C9RVK4 A0A3L8DE08 A0A0V0GA44 T1HIG2 A0A069DYT0 A0A023FWI7 A0A2R5LJQ5 A0A224XEF2 B4PY70 V3ZKD4 A0A151I0X6 Q9W552 B4I9J7 A0A0L7R868
A0A194RJK5 A0A212FA24 A0A1L8DVC7 A0A336M094 A0A336MUK6 A0A0K8TSB1 U5EWK8 D2A583 A0A0T6BD35 A0A1I8MNP2 A0A2M4AFJ4 A0A1Q3F910 A0A1Q3F8S6 A0A2M3Z479 A0A2M4AFT7 A0A2M4BHM7 A0A2M4BHJ9 A0A2M4BHS7 W5J8V2 D3TNJ9 A0A0L0C435 A0A2M3Z426 A0A1S4G1K5 A0A182XP88 A0A067QYJ0 Q7Q5Y3 A0A2J7Q1J4 A0A0A1WRR9 A0A1A9WA17 A0A182Y2R6 A0A182KNZ4 A0A1A9XM10 A0A034WM28 A0A182I1D8 A0A1Y9HDZ0 A0A182UPZ1 A0A182FEI9 A0A182GF45 A0A1A9Z3Y4 A0A2M3Z558 W8BR63 A0A1W4X737 A0A226CXH6 A0A182JPN5 A0A182LXX7 A0A1A9VWW5 A0A1E1XTV5 A0A182IL17 A0A1J1J444 A0A084WEQ8 A0A182PDS1 A0A023GLK5 G3MM37 A0A1E1X9Z7 A0A0K8W0J3 A0A0M4FAP9 A0A023FIT6 Q16KJ7 A0A1B6L663 A0A1B6FGW5 B7PQ07 V5H0U0 V9ILB7 A0A088A5E3 A0A1B6H6C5 A0A1S3HY72 A0A2A3E6Q0 A0A0M9A5I2 A0A0C9RVK4 A0A3L8DE08 A0A0V0GA44 T1HIG2 A0A069DYT0 A0A023FWI7 A0A2R5LJQ5 A0A224XEF2 B4PY70 V3ZKD4 A0A151I0X6 Q9W552 B4I9J7 A0A0L7R868
PDB
2R51
E-value=2.04418e-131,
Score=1202
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Membrane
Membrane
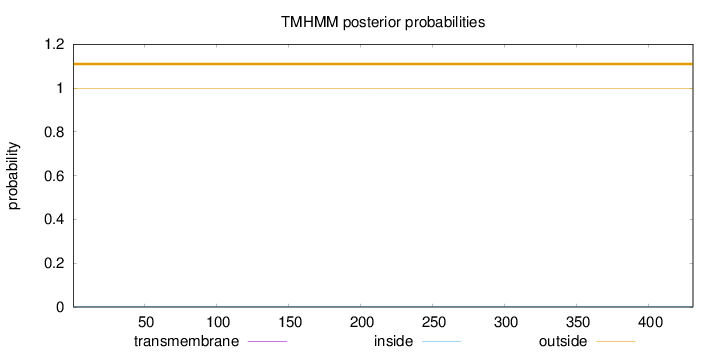
Length:
431
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0011
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00092
outside
1 - 431
Population Genetic Test Statistics
Pi
203.410464
Theta
177.033701
Tajima's D
0.546991
CLR
0.039664
CSRT
0.529173541322934
Interpretation
Uncertain
