Pre Gene Modal
BGIBMGA000119
Annotation
PREDICTED:_porphobilinogen_deaminase_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 1.996
Sequence
CDS
ATGGACGGTGAAACGAAACTAAGCATTCACGTCGGCTCCAGGAAGAGCGAGCTAGCACTTGTACAAACGAATTTCGTTATCGATAGCTTAAAAAGAAATTACCCGGAAAAGGAATTTAAAATTGTGACCATGACAACGCTTGGCGACAGGGTGCTAGATCAGCCTCTACCAAAAATAGGAGAAAAGTCACTTTTCACAAAAGACTTAGAAGATGCACTGATGAGTAAAAATGTAGATTTTGTTGTGCATTCACTGAAAGACTTGCCAACAACATTGCCGGACGGATTAGTCATTGGCGCTGTCTTTGAAAGAGAAGACCCTCGTGATGCTTTGGTACTGAACGAAAAATTTAAGGAGTACTCATTGTCCACCTTACCTTCAGGATCAGTAATAGGAACATCATCACTGAGACGAACGGCTCAGCTAAACGGAAACTATCCCCAGTTGAAGGTGGTGGATGTGAGAGGCAATCTTAATACCAGGTTGAGGAAATTGGATGACGAAGATGGAAAATATTCAGCCCTTATACTGGCAAGTGCTGGGTTGAGTAGAATGGGATGGGGCAACAGAATGTCAAAGGTACTGCCGTGCTCGGAGATGTTATATGCGGTGGGTCAGGGCGCGTTGGCGGTGGAATGTCGCGCTGACAATGAAGAAGTCCTGGCGATGTTGGCTCCGTTCAACCACCCTGAAACTTACTGCAGGGTCTTGGCTGAGAGGAGTTTCCTAAAAACTTTAGGTGGTGGATGCAGCGCTCCGGTAGGTATATCAACAAAACTAAAACAAAACGATAATGTATACAAGCTCACTTTGACCGGAGCCGTCTGGAGTCTAGACGGTTCGACCAAACTCGAGGAAACGTTGGACCAAACGTTTGGGCAAATACGTAAAACCGTTAAACACAAACTCAGTCCGACGGAAGAAGCGAGCAGCAAAAAAATTAAAACCGACAAAACAGTTGAAGCCGACAATGAAATCGCCGTACTTAATCGACGGATCACCGACAAGACTGGTGATTTGGGCTGCGAAGATTTGGCGCCAAATGTCGCTGACAGACGCTGCTTCTGCGGTTTGACGACCAACGTCAATATGCCAATTGACGTTGTCATCAAATGTGATAATTTAGGTAGGGACTTGGCGAATTCGTTAATCGATAAAGGTGCTTTAGAGGTGATGAAGATATCACATGACATAATCAGAAGTTCGATTGGTAAAAAATCATGA
Protein
MDGETKLSIHVGSRKSELALVQTNFVIDSLKRNYPEKEFKIVTMTTLGDRVLDQPLPKIGEKSLFTKDLEDALMSKNVDFVVHSLKDLPTTLPDGLVIGAVFEREDPRDALVLNEKFKEYSLSTLPSGSVIGTSSLRRTAQLNGNYPQLKVVDVRGNLNTRLRKLDDEDGKYSALILASAGLSRMGWGNRMSKVLPCSEMLYAVGQGALAVECRADNEEVLAMLAPFNHPETYCRVLAERSFLKTLGGGCSAPVGISTKLKQNDNVYKLTLTGAVWSLDGSTKLEETLDQTFGQIRKTVKHKLSPTEEASSKKIKTDKTVEADNEIAVLNRRITDKTGDLGCEDLAPNVADRRCFCGLTTNVNMPIDVVIKCDNLGRDLANSLIDKGALEVMKISHDIIRSSIGKKS
Summary
Uniprot
H9IS92
A0A3S2NDX7
A0A194QH33
A0A2A4JUN9
A0A2H1WSS2
A0A212FA34
+ More
A0A1B6LZS6 A0A1B6EJB0 V5GX26 A0A1B6D9V3 J9K6V0 A0A2H8TQ12 A0A1Y1LL53 A0A2S2RAS4 W8B9N7 A0A1I8MZD9 T1PBM0 B4HVS3 B3M516 A0A2J7RE79 A0A034WJ22 A0A1W4W5Y3 A0A0M5J056 A0A0K8WC50 Q9W0G4 B4PD78 A0A0A1XIU8 B4QM08 A0A232F4J1 A0A1I8Q077 A0A1Q3G1Z3 A0A1Q3G1K5 A0A3L8DV68 B3NB99 B4KY05 B4N3A3 A0A1Q3G1I6 A0A1J1J529 A0A0K8TSR8 A0A3B0JYY7 U5EPV1 B4LG64 Q29EW7 A0A1B0FG25 A0A2M4A896 A0A088ALA1 A0A2A3ETM6 A0A182QUV5 B4IXG6 A0A1B0ANK3 A0A2M4A503 A0A2M4A5E0 A0A182JS32 A0A2M4CIQ7 A0A2M4BGY7 A0A2M4BGY4 A0A2M3Z6X0 A0A084WLV2 A0A0A9X341 A0A2M3Z6B8 A0A2M3Z615 A0A2M3Z640 A0A2M3Z8R1 A0A182NSX9 A0A2M4CR78 A0A182YAU3 A0A2M4CRD1 A0A182RD70 A0A182MF69 B4H5M2 A0A2R7VY17 A0A182WUD9 A0A1S4H6K0 A0A182UV24 A0A182KKT1 A0A182HW36 A0A182WDS5 A0A1A9ZBT7 A0A1S3D3D3 F4X8M7 A0A0P4VJZ0 E2A1J7 T1HIA9 A0A0P5P2Q2 A0A162SUT7 A0A182FTL6
A0A1B6LZS6 A0A1B6EJB0 V5GX26 A0A1B6D9V3 J9K6V0 A0A2H8TQ12 A0A1Y1LL53 A0A2S2RAS4 W8B9N7 A0A1I8MZD9 T1PBM0 B4HVS3 B3M516 A0A2J7RE79 A0A034WJ22 A0A1W4W5Y3 A0A0M5J056 A0A0K8WC50 Q9W0G4 B4PD78 A0A0A1XIU8 B4QM08 A0A232F4J1 A0A1I8Q077 A0A1Q3G1Z3 A0A1Q3G1K5 A0A3L8DV68 B3NB99 B4KY05 B4N3A3 A0A1Q3G1I6 A0A1J1J529 A0A0K8TSR8 A0A3B0JYY7 U5EPV1 B4LG64 Q29EW7 A0A1B0FG25 A0A2M4A896 A0A088ALA1 A0A2A3ETM6 A0A182QUV5 B4IXG6 A0A1B0ANK3 A0A2M4A503 A0A2M4A5E0 A0A182JS32 A0A2M4CIQ7 A0A2M4BGY7 A0A2M4BGY4 A0A2M3Z6X0 A0A084WLV2 A0A0A9X341 A0A2M3Z6B8 A0A2M3Z615 A0A2M3Z640 A0A2M3Z8R1 A0A182NSX9 A0A2M4CR78 A0A182YAU3 A0A2M4CRD1 A0A182RD70 A0A182MF69 B4H5M2 A0A2R7VY17 A0A182WUD9 A0A1S4H6K0 A0A182UV24 A0A182KKT1 A0A182HW36 A0A182WDS5 A0A1A9ZBT7 A0A1S3D3D3 F4X8M7 A0A0P4VJZ0 E2A1J7 T1HIA9 A0A0P5P2Q2 A0A162SUT7 A0A182FTL6
Pubmed
19121390
26354079
22118469
28004739
24495485
25315136
+ More
17994087 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25830018 22936249 28648823 30249741 26369729 15632085 24438588 25401762 26823975 25244985 12364791 20966253 21719571 27129103 20798317
17994087 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25830018 22936249 28648823 30249741 26369729 15632085 24438588 25401762 26823975 25244985 12364791 20966253 21719571 27129103 20798317
EMBL
BABH01024104
BABH01024105
BABH01024106
RSAL01000179
RVE44968.1
KQ458859
+ More
KPJ04853.1 NWSH01000581 PCG75519.1 ODYU01010731 SOQ56022.1 AGBW02009528 OWR50583.1 GEBQ01010788 JAT29189.1 GECZ01031750 JAS38019.1 GALX01003608 JAB64858.1 GEDC01014898 JAS22400.1 ABLF02014710 GFXV01004482 MBW16287.1 GEZM01054541 JAV73568.1 GGMS01017954 MBY87157.1 GAMC01016669 JAB89886.1 KA646161 AFP60790.1 CH480817 EDW50038.1 CH902618 EDV39495.1 NEVH01005277 PNF39142.1 GAKP01003376 GAKP01003374 JAC55576.1 CP012525 ALC42998.1 GDHF01003675 JAI48639.1 AE014296 BT023907 BT050461 AAF47484.1 ABA81841.1 ACJ13168.1 CM000159 EDW92826.1 GBXI01003834 JAD10458.1 CM000363 CM002912 EDX08794.1 KMY96806.1 NNAY01001035 OXU25400.1 GFDL01001232 JAV33813.1 GFDL01001360 JAV33685.1 QOIP01000004 RLU24103.1 CH954178 EDV50152.1 CH933809 EDW18707.1 CH964062 EDW78842.1 GFDL01001406 JAV33639.1 CVRI01000072 CRL07515.1 GDAI01000196 JAI17407.1 OUUW01000012 SPP87264.1 GANO01003557 JAB56314.1 CH940647 EDW69372.1 CH379070 EAL29942.2 CCAG010003862 GGFK01003639 MBW36960.1 KZ288189 PBC34566.1 AXCN02002132 AXCN02002133 CH916366 EDV96403.1 JXJN01000881 GGFK01002556 MBW35877.1 GGFK01002527 MBW35848.1 GGFL01001048 MBW65226.1 GGFJ01003179 MBW52320.1 GGFJ01003178 MBW52319.1 GGFM01003525 MBW24276.1 ATLV01024293 ATLV01024294 KE525351 KFB51196.1 GBHO01032079 GBRD01001003 GDHC01017752 JAG11525.1 JAG64818.1 JAQ00877.1 GGFM01003294 MBW24045.1 GGFM01003199 MBW23950.1 GGFM01003235 MBW23986.1 GGFM01004134 MBW24885.1 GGFL01003658 MBW67836.1 GGFL01003657 MBW67835.1 AXCM01016885 CH479211 EDW33074.1 KK854163 PTY12396.1 AAAB01008816 APCN01001462 GL888932 EGI57296.1 GDKW01002023 JAI54572.1 GL435766 EFN72670.1 ACPB03006711 GDIQ01155326 GDIP01060317 JAK96399.1 JAM43398.1 LRGB01000024 KZS21644.1
KPJ04853.1 NWSH01000581 PCG75519.1 ODYU01010731 SOQ56022.1 AGBW02009528 OWR50583.1 GEBQ01010788 JAT29189.1 GECZ01031750 JAS38019.1 GALX01003608 JAB64858.1 GEDC01014898 JAS22400.1 ABLF02014710 GFXV01004482 MBW16287.1 GEZM01054541 JAV73568.1 GGMS01017954 MBY87157.1 GAMC01016669 JAB89886.1 KA646161 AFP60790.1 CH480817 EDW50038.1 CH902618 EDV39495.1 NEVH01005277 PNF39142.1 GAKP01003376 GAKP01003374 JAC55576.1 CP012525 ALC42998.1 GDHF01003675 JAI48639.1 AE014296 BT023907 BT050461 AAF47484.1 ABA81841.1 ACJ13168.1 CM000159 EDW92826.1 GBXI01003834 JAD10458.1 CM000363 CM002912 EDX08794.1 KMY96806.1 NNAY01001035 OXU25400.1 GFDL01001232 JAV33813.1 GFDL01001360 JAV33685.1 QOIP01000004 RLU24103.1 CH954178 EDV50152.1 CH933809 EDW18707.1 CH964062 EDW78842.1 GFDL01001406 JAV33639.1 CVRI01000072 CRL07515.1 GDAI01000196 JAI17407.1 OUUW01000012 SPP87264.1 GANO01003557 JAB56314.1 CH940647 EDW69372.1 CH379070 EAL29942.2 CCAG010003862 GGFK01003639 MBW36960.1 KZ288189 PBC34566.1 AXCN02002132 AXCN02002133 CH916366 EDV96403.1 JXJN01000881 GGFK01002556 MBW35877.1 GGFK01002527 MBW35848.1 GGFL01001048 MBW65226.1 GGFJ01003179 MBW52320.1 GGFJ01003178 MBW52319.1 GGFM01003525 MBW24276.1 ATLV01024293 ATLV01024294 KE525351 KFB51196.1 GBHO01032079 GBRD01001003 GDHC01017752 JAG11525.1 JAG64818.1 JAQ00877.1 GGFM01003294 MBW24045.1 GGFM01003199 MBW23950.1 GGFM01003235 MBW23986.1 GGFM01004134 MBW24885.1 GGFL01003658 MBW67836.1 GGFL01003657 MBW67835.1 AXCM01016885 CH479211 EDW33074.1 KK854163 PTY12396.1 AAAB01008816 APCN01001462 GL888932 EGI57296.1 GDKW01002023 JAI54572.1 GL435766 EFN72670.1 ACPB03006711 GDIQ01155326 GDIP01060317 JAK96399.1 JAM43398.1 LRGB01000024 KZS21644.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000007151
UP000007819
+ More
UP000095301 UP000001292 UP000007801 UP000235965 UP000192221 UP000092553 UP000000803 UP000002282 UP000000304 UP000215335 UP000095300 UP000279307 UP000008711 UP000009192 UP000007798 UP000183832 UP000268350 UP000008792 UP000001819 UP000092444 UP000005203 UP000242457 UP000075886 UP000001070 UP000092460 UP000075881 UP000030765 UP000075884 UP000076408 UP000075900 UP000075883 UP000008744 UP000076407 UP000075903 UP000075882 UP000075840 UP000075920 UP000092445 UP000079169 UP000007755 UP000000311 UP000015103 UP000076858 UP000069272
UP000095301 UP000001292 UP000007801 UP000235965 UP000192221 UP000092553 UP000000803 UP000002282 UP000000304 UP000215335 UP000095300 UP000279307 UP000008711 UP000009192 UP000007798 UP000183832 UP000268350 UP000008792 UP000001819 UP000092444 UP000005203 UP000242457 UP000075886 UP000001070 UP000092460 UP000075881 UP000030765 UP000075884 UP000076408 UP000075900 UP000075883 UP000008744 UP000076407 UP000075903 UP000075882 UP000075840 UP000075920 UP000092445 UP000079169 UP000007755 UP000000311 UP000015103 UP000076858 UP000069272
Interpro
SUPFAM
SSF54782
SSF54782
Gene 3D
ProteinModelPortal
H9IS92
A0A3S2NDX7
A0A194QH33
A0A2A4JUN9
A0A2H1WSS2
A0A212FA34
+ More
A0A1B6LZS6 A0A1B6EJB0 V5GX26 A0A1B6D9V3 J9K6V0 A0A2H8TQ12 A0A1Y1LL53 A0A2S2RAS4 W8B9N7 A0A1I8MZD9 T1PBM0 B4HVS3 B3M516 A0A2J7RE79 A0A034WJ22 A0A1W4W5Y3 A0A0M5J056 A0A0K8WC50 Q9W0G4 B4PD78 A0A0A1XIU8 B4QM08 A0A232F4J1 A0A1I8Q077 A0A1Q3G1Z3 A0A1Q3G1K5 A0A3L8DV68 B3NB99 B4KY05 B4N3A3 A0A1Q3G1I6 A0A1J1J529 A0A0K8TSR8 A0A3B0JYY7 U5EPV1 B4LG64 Q29EW7 A0A1B0FG25 A0A2M4A896 A0A088ALA1 A0A2A3ETM6 A0A182QUV5 B4IXG6 A0A1B0ANK3 A0A2M4A503 A0A2M4A5E0 A0A182JS32 A0A2M4CIQ7 A0A2M4BGY7 A0A2M4BGY4 A0A2M3Z6X0 A0A084WLV2 A0A0A9X341 A0A2M3Z6B8 A0A2M3Z615 A0A2M3Z640 A0A2M3Z8R1 A0A182NSX9 A0A2M4CR78 A0A182YAU3 A0A2M4CRD1 A0A182RD70 A0A182MF69 B4H5M2 A0A2R7VY17 A0A182WUD9 A0A1S4H6K0 A0A182UV24 A0A182KKT1 A0A182HW36 A0A182WDS5 A0A1A9ZBT7 A0A1S3D3D3 F4X8M7 A0A0P4VJZ0 E2A1J7 T1HIA9 A0A0P5P2Q2 A0A162SUT7 A0A182FTL6
A0A1B6LZS6 A0A1B6EJB0 V5GX26 A0A1B6D9V3 J9K6V0 A0A2H8TQ12 A0A1Y1LL53 A0A2S2RAS4 W8B9N7 A0A1I8MZD9 T1PBM0 B4HVS3 B3M516 A0A2J7RE79 A0A034WJ22 A0A1W4W5Y3 A0A0M5J056 A0A0K8WC50 Q9W0G4 B4PD78 A0A0A1XIU8 B4QM08 A0A232F4J1 A0A1I8Q077 A0A1Q3G1Z3 A0A1Q3G1K5 A0A3L8DV68 B3NB99 B4KY05 B4N3A3 A0A1Q3G1I6 A0A1J1J529 A0A0K8TSR8 A0A3B0JYY7 U5EPV1 B4LG64 Q29EW7 A0A1B0FG25 A0A2M4A896 A0A088ALA1 A0A2A3ETM6 A0A182QUV5 B4IXG6 A0A1B0ANK3 A0A2M4A503 A0A2M4A5E0 A0A182JS32 A0A2M4CIQ7 A0A2M4BGY7 A0A2M4BGY4 A0A2M3Z6X0 A0A084WLV2 A0A0A9X341 A0A2M3Z6B8 A0A2M3Z615 A0A2M3Z640 A0A2M3Z8R1 A0A182NSX9 A0A2M4CR78 A0A182YAU3 A0A2M4CRD1 A0A182RD70 A0A182MF69 B4H5M2 A0A2R7VY17 A0A182WUD9 A0A1S4H6K0 A0A182UV24 A0A182KKT1 A0A182HW36 A0A182WDS5 A0A1A9ZBT7 A0A1S3D3D3 F4X8M7 A0A0P4VJZ0 E2A1J7 T1HIA9 A0A0P5P2Q2 A0A162SUT7 A0A182FTL6
PDB
5M7F
E-value=7.91539e-85,
Score=800
Ontologies
KEGG
PATHWAY
GO
PANTHER
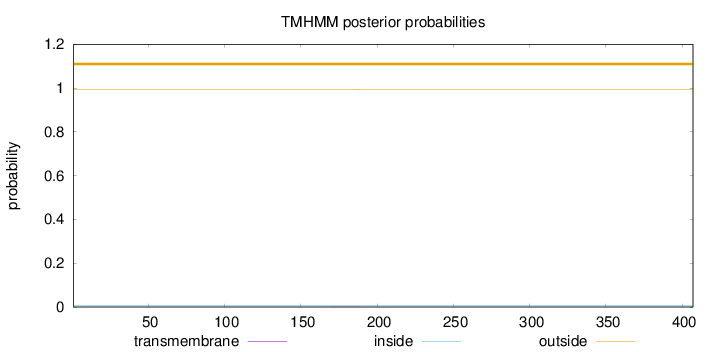
Topology
Length:
407
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00713
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00702
outside
1 - 407
Population Genetic Test Statistics
Pi
291.409772
Theta
179.052906
Tajima's D
2.199483
CLR
0.018732
CSRT
0.914454277286136
Interpretation
Uncertain
