Gene
KWMTBOMO14404
Pre Gene Modal
BGIBMGA000110
Annotation
PREDICTED:_serologically_defined_colon_cancer_antigen_8_homolog_isoform_X2_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.715
Sequence
CDS
ATGCCATCACATAATGCAAAAGCTCAAAAGCTTGACTTGGACCGTCAGCGGCGCGAGGAGAACGATCTCAAACGAGAACTGACCTTGAAGAACTCCGCCGTTGAGGAACTGAAGATGGAACTCAAGAATAAAACTGCATCTCTCCAGGCTGATCTGTCTCAGGCTCACGCCGAGAAGGCTTCATTGGAGGAGGAGCTGGCAAGCGCGCGGCTGGCCTTAGAGCGACACCAGCGCCAGACCAAACACGAAACTAATCGACTTAATTCAGAGATCCAGTCCCTTCGTCAGCGTCTGGACCGAGCTGACGCGGACCTGGTACACTCGCGCCGCGAGAACCTGCGTCTCTCTGAGCAGATCTCCAATCTCGAAAAAGAGATTAACTTGAAGAGCTTGAGCCCGATCTCTTCAGAGAAGAACAAGAAGGAAAAGGAGCTGTCTACCATGCTGGAGTCCATTGACAACAAGCACGGTGCGTACACCAGCACCACTATACAGACGTAG
Protein
MPSHNAKAQKLDLDRQRREENDLKRELTLKNSAVEELKMELKNKTASLQADLSQAHAEKASLEEELASARLALERHQRQTKHETNRLNSEIQSLRQRLDRADADLVHSRRENLRLSEQISNLEKEINLKSLSPISSEKNKKEKELSTMLESIDNKHGAYTSTTIQT
Summary
Uniprot
A0A3S2NPB7
A0A194RIH2
A0A212FHQ5
A0A2H1WXQ4
A0A194QMZ5
A0A0N1IIF3
+ More
Q16JS9 A0A182HD36 U4UA84 N6UDW2 W5JM21 A0A1S4FYJ5 B0XDC5 A0A1B0F0M4 A0A182Q2P5 A0A1B0CL97 A0A182PD93 A0A084WNK5 A0A182KV38 A0A182RW78 A0A182WE48 A0A182LUM2 A0A182Y9N1 A0A182WXW5 A0A182I5Y6 A0A182UME1 A0A2Y9D2E5 U5EUK6 A0A182FVG3 A0A182IKQ7 A0A182JX89 A0A182UI48 A0A182S8N7 A0A182N2N5 A0A1B6DK32 A0A0L0CEB9 A0A067R4F6 F4WKT9 A0A2J7QB27 A0A1W4VHW4 A0A139WKF1 A0A139WKJ6 A0A1W4VHL7 A0A1W4VUF0 A0A1W4VV37 A0A1W4VV49 A0A1I8PNM9 A0A1I8PNQ2 A0A2J7QB13 E2AJ52 A0A151JVJ3 A0A195CN41 A0A195BJH8 A0A1I8PNQ7 A0A1I8PNT3 A0A158P3Y4 A0A1I8PNS8 A0A1I8PNN4 A0A0R1E385 A0A0R1E955 Q6NR11 E2BGG7 A0A146L9C7 A0A0B4LIA2 B4PRI8 A0A0K8SIA7 A0A0R1E3A2 A0A0P8Y0L7 B4JT52 A0A0P9ASD2 A0A0A9WSU1 A0A0B4LHH2 Q9VF13 A0A0J7KDD2 Q9VF12 A0A0A9WU78 B3M393 A0A0B4LH69 A0A2J7QB19 A0A0B4LHA1 A0A1J1ISH3 B3P3X1 B4NBA2 A0A151X116 A0A1I8M229 B4QXT1 B4K5Q4 B4HLG5 A0A026WX91 A0A151J8U2 A0A0K8T5B5 A0A3L8D8L1 A0A0A9WA39 A0A146L8Z3 B4LX92 A0A0A9WIE3 A0A2P8XQS2 A0A1B6JHP8
Q16JS9 A0A182HD36 U4UA84 N6UDW2 W5JM21 A0A1S4FYJ5 B0XDC5 A0A1B0F0M4 A0A182Q2P5 A0A1B0CL97 A0A182PD93 A0A084WNK5 A0A182KV38 A0A182RW78 A0A182WE48 A0A182LUM2 A0A182Y9N1 A0A182WXW5 A0A182I5Y6 A0A182UME1 A0A2Y9D2E5 U5EUK6 A0A182FVG3 A0A182IKQ7 A0A182JX89 A0A182UI48 A0A182S8N7 A0A182N2N5 A0A1B6DK32 A0A0L0CEB9 A0A067R4F6 F4WKT9 A0A2J7QB27 A0A1W4VHW4 A0A139WKF1 A0A139WKJ6 A0A1W4VHL7 A0A1W4VUF0 A0A1W4VV37 A0A1W4VV49 A0A1I8PNM9 A0A1I8PNQ2 A0A2J7QB13 E2AJ52 A0A151JVJ3 A0A195CN41 A0A195BJH8 A0A1I8PNQ7 A0A1I8PNT3 A0A158P3Y4 A0A1I8PNS8 A0A1I8PNN4 A0A0R1E385 A0A0R1E955 Q6NR11 E2BGG7 A0A146L9C7 A0A0B4LIA2 B4PRI8 A0A0K8SIA7 A0A0R1E3A2 A0A0P8Y0L7 B4JT52 A0A0P9ASD2 A0A0A9WSU1 A0A0B4LHH2 Q9VF13 A0A0J7KDD2 Q9VF12 A0A0A9WU78 B3M393 A0A0B4LH69 A0A2J7QB19 A0A0B4LHA1 A0A1J1ISH3 B3P3X1 B4NBA2 A0A151X116 A0A1I8M229 B4QXT1 B4K5Q4 B4HLG5 A0A026WX91 A0A151J8U2 A0A0K8T5B5 A0A3L8D8L1 A0A0A9WA39 A0A146L8Z3 B4LX92 A0A0A9WIE3 A0A2P8XQS2 A0A1B6JHP8
Pubmed
26354079
22118469
17510324
26483478
23537049
20920257
+ More
23761445 24438588 20966253 25244985 26108605 24845553 21719571 18362917 19820115 20798317 21347285 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26823975 25401762 25315136 24508170 30249741 29403074
23761445 24438588 20966253 25244985 26108605 24845553 21719571 18362917 19820115 20798317 21347285 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26823975 25401762 25315136 24508170 30249741 29403074
EMBL
RSAL01000179
RVE44965.1
KQ460124
KPJ17623.1
AGBW02008471
OWR53269.1
+ More
ODYU01011436 SOQ57174.1 KQ458859 KPJ04856.1 LADJ01066008 KPJ21679.1 CH477994 EAT34532.1 JXUM01034368 JXUM01034369 JXUM01034370 KQ561021 KXJ79997.1 KB632195 ERL89952.1 APGK01026643 KB740635 ENN79895.1 ADMH02001177 ETN63829.1 DS232752 EDS45403.1 AJVK01007785 AXCN02001080 AJWK01017052 AJWK01017053 AJWK01017054 ATLV01024620 KE525353 KFB51799.1 AXCM01004280 APCN01003537 GANO01003837 JAB56034.1 GEDC01011244 JAS26054.1 JRES01000623 KNC29824.1 KK852984 KDR12857.1 GL888206 EGI65181.1 NEVH01016304 PNF25779.1 KQ971326 KYB28459.1 KYB28460.1 PNF25776.1 GL439967 EFN66492.1 KQ981701 KYN37496.1 KQ977513 KYN02166.1 KQ976453 KYM85341.1 ADTU01008610 CM000160 KRK03718.1 KRK03719.1 BT010273 AE014297 AAQ23591.1 ABW08684.1 GL448181 EFN85216.1 GDHC01014300 JAQ04329.1 AHN57347.1 EDW97388.2 GBRD01012815 JAG53011.1 KRK03720.1 CH902617 KPU80312.1 CH916373 EDV94942.1 KPU80313.1 GBHO01035684 JAG07920.1 AHN57345.1 AAF55249.2 LBMM01009012 KMQ88463.1 AAF55250.2 GBHO01032280 JAG11324.1 EDV43554.1 AHN57343.1 AHN57344.1 PNF25780.1 AHN57346.1 CVRI01000059 CRL03187.1 CH954181 EDV49077.1 CH964232 EDW81066.2 KQ982612 KYQ53869.1 CM000364 EDX12755.1 CH933806 EDW15116.2 CH480815 EDW41985.1 KK107078 EZA60458.1 KQ979480 KYN21282.1 GBRD01005099 JAG60722.1 QOIP01000011 RLU16626.1 GBHO01038995 JAG04609.1 GDHC01014554 JAQ04075.1 CH940650 EDW66744.2 GBHO01039009 JAG04595.1 PYGN01001512 PSN34350.1 GECU01008998 JAS98708.1
ODYU01011436 SOQ57174.1 KQ458859 KPJ04856.1 LADJ01066008 KPJ21679.1 CH477994 EAT34532.1 JXUM01034368 JXUM01034369 JXUM01034370 KQ561021 KXJ79997.1 KB632195 ERL89952.1 APGK01026643 KB740635 ENN79895.1 ADMH02001177 ETN63829.1 DS232752 EDS45403.1 AJVK01007785 AXCN02001080 AJWK01017052 AJWK01017053 AJWK01017054 ATLV01024620 KE525353 KFB51799.1 AXCM01004280 APCN01003537 GANO01003837 JAB56034.1 GEDC01011244 JAS26054.1 JRES01000623 KNC29824.1 KK852984 KDR12857.1 GL888206 EGI65181.1 NEVH01016304 PNF25779.1 KQ971326 KYB28459.1 KYB28460.1 PNF25776.1 GL439967 EFN66492.1 KQ981701 KYN37496.1 KQ977513 KYN02166.1 KQ976453 KYM85341.1 ADTU01008610 CM000160 KRK03718.1 KRK03719.1 BT010273 AE014297 AAQ23591.1 ABW08684.1 GL448181 EFN85216.1 GDHC01014300 JAQ04329.1 AHN57347.1 EDW97388.2 GBRD01012815 JAG53011.1 KRK03720.1 CH902617 KPU80312.1 CH916373 EDV94942.1 KPU80313.1 GBHO01035684 JAG07920.1 AHN57345.1 AAF55249.2 LBMM01009012 KMQ88463.1 AAF55250.2 GBHO01032280 JAG11324.1 EDV43554.1 AHN57343.1 AHN57344.1 PNF25780.1 AHN57346.1 CVRI01000059 CRL03187.1 CH954181 EDV49077.1 CH964232 EDW81066.2 KQ982612 KYQ53869.1 CM000364 EDX12755.1 CH933806 EDW15116.2 CH480815 EDW41985.1 KK107078 EZA60458.1 KQ979480 KYN21282.1 GBRD01005099 JAG60722.1 QOIP01000011 RLU16626.1 GBHO01038995 JAG04609.1 GDHC01014554 JAQ04075.1 CH940650 EDW66744.2 GBHO01039009 JAG04595.1 PYGN01001512 PSN34350.1 GECU01008998 JAS98708.1
Proteomes
UP000283053
UP000053240
UP000007151
UP000053268
UP000008820
UP000069940
+ More
UP000249989 UP000030742 UP000019118 UP000000673 UP000002320 UP000092462 UP000075886 UP000092461 UP000075885 UP000030765 UP000075882 UP000075900 UP000075920 UP000075883 UP000076408 UP000076407 UP000075840 UP000075903 UP000069272 UP000075880 UP000075881 UP000075902 UP000075901 UP000075884 UP000037069 UP000027135 UP000007755 UP000235965 UP000192221 UP000007266 UP000095300 UP000000311 UP000078541 UP000078542 UP000078540 UP000005205 UP000002282 UP000000803 UP000008237 UP000007801 UP000001070 UP000036403 UP000183832 UP000008711 UP000007798 UP000075809 UP000095301 UP000000304 UP000009192 UP000001292 UP000053097 UP000078492 UP000279307 UP000008792 UP000245037
UP000249989 UP000030742 UP000019118 UP000000673 UP000002320 UP000092462 UP000075886 UP000092461 UP000075885 UP000030765 UP000075882 UP000075900 UP000075920 UP000075883 UP000076408 UP000076407 UP000075840 UP000075903 UP000069272 UP000075880 UP000075881 UP000075902 UP000075901 UP000075884 UP000037069 UP000027135 UP000007755 UP000235965 UP000192221 UP000007266 UP000095300 UP000000311 UP000078541 UP000078542 UP000078540 UP000005205 UP000002282 UP000000803 UP000008237 UP000007801 UP000001070 UP000036403 UP000183832 UP000008711 UP000007798 UP000075809 UP000095301 UP000000304 UP000009192 UP000001292 UP000053097 UP000078492 UP000279307 UP000008792 UP000245037
ProteinModelPortal
A0A3S2NPB7
A0A194RIH2
A0A212FHQ5
A0A2H1WXQ4
A0A194QMZ5
A0A0N1IIF3
+ More
Q16JS9 A0A182HD36 U4UA84 N6UDW2 W5JM21 A0A1S4FYJ5 B0XDC5 A0A1B0F0M4 A0A182Q2P5 A0A1B0CL97 A0A182PD93 A0A084WNK5 A0A182KV38 A0A182RW78 A0A182WE48 A0A182LUM2 A0A182Y9N1 A0A182WXW5 A0A182I5Y6 A0A182UME1 A0A2Y9D2E5 U5EUK6 A0A182FVG3 A0A182IKQ7 A0A182JX89 A0A182UI48 A0A182S8N7 A0A182N2N5 A0A1B6DK32 A0A0L0CEB9 A0A067R4F6 F4WKT9 A0A2J7QB27 A0A1W4VHW4 A0A139WKF1 A0A139WKJ6 A0A1W4VHL7 A0A1W4VUF0 A0A1W4VV37 A0A1W4VV49 A0A1I8PNM9 A0A1I8PNQ2 A0A2J7QB13 E2AJ52 A0A151JVJ3 A0A195CN41 A0A195BJH8 A0A1I8PNQ7 A0A1I8PNT3 A0A158P3Y4 A0A1I8PNS8 A0A1I8PNN4 A0A0R1E385 A0A0R1E955 Q6NR11 E2BGG7 A0A146L9C7 A0A0B4LIA2 B4PRI8 A0A0K8SIA7 A0A0R1E3A2 A0A0P8Y0L7 B4JT52 A0A0P9ASD2 A0A0A9WSU1 A0A0B4LHH2 Q9VF13 A0A0J7KDD2 Q9VF12 A0A0A9WU78 B3M393 A0A0B4LH69 A0A2J7QB19 A0A0B4LHA1 A0A1J1ISH3 B3P3X1 B4NBA2 A0A151X116 A0A1I8M229 B4QXT1 B4K5Q4 B4HLG5 A0A026WX91 A0A151J8U2 A0A0K8T5B5 A0A3L8D8L1 A0A0A9WA39 A0A146L8Z3 B4LX92 A0A0A9WIE3 A0A2P8XQS2 A0A1B6JHP8
Q16JS9 A0A182HD36 U4UA84 N6UDW2 W5JM21 A0A1S4FYJ5 B0XDC5 A0A1B0F0M4 A0A182Q2P5 A0A1B0CL97 A0A182PD93 A0A084WNK5 A0A182KV38 A0A182RW78 A0A182WE48 A0A182LUM2 A0A182Y9N1 A0A182WXW5 A0A182I5Y6 A0A182UME1 A0A2Y9D2E5 U5EUK6 A0A182FVG3 A0A182IKQ7 A0A182JX89 A0A182UI48 A0A182S8N7 A0A182N2N5 A0A1B6DK32 A0A0L0CEB9 A0A067R4F6 F4WKT9 A0A2J7QB27 A0A1W4VHW4 A0A139WKF1 A0A139WKJ6 A0A1W4VHL7 A0A1W4VUF0 A0A1W4VV37 A0A1W4VV49 A0A1I8PNM9 A0A1I8PNQ2 A0A2J7QB13 E2AJ52 A0A151JVJ3 A0A195CN41 A0A195BJH8 A0A1I8PNQ7 A0A1I8PNT3 A0A158P3Y4 A0A1I8PNS8 A0A1I8PNN4 A0A0R1E385 A0A0R1E955 Q6NR11 E2BGG7 A0A146L9C7 A0A0B4LIA2 B4PRI8 A0A0K8SIA7 A0A0R1E3A2 A0A0P8Y0L7 B4JT52 A0A0P9ASD2 A0A0A9WSU1 A0A0B4LHH2 Q9VF13 A0A0J7KDD2 Q9VF12 A0A0A9WU78 B3M393 A0A0B4LH69 A0A2J7QB19 A0A0B4LHA1 A0A1J1ISH3 B3P3X1 B4NBA2 A0A151X116 A0A1I8M229 B4QXT1 B4K5Q4 B4HLG5 A0A026WX91 A0A151J8U2 A0A0K8T5B5 A0A3L8D8L1 A0A0A9WA39 A0A146L8Z3 B4LX92 A0A0A9WIE3 A0A2P8XQS2 A0A1B6JHP8
Ontologies
PANTHER
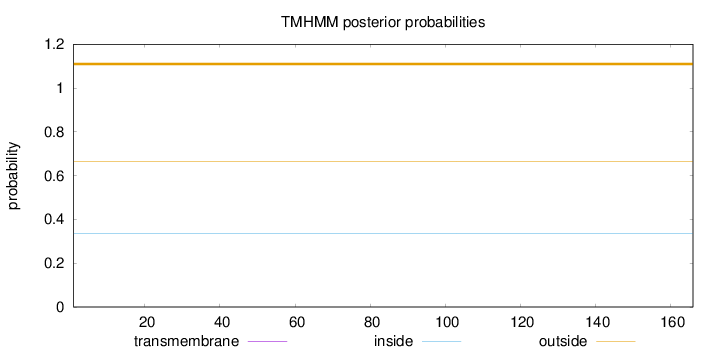
Topology
Length:
166
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.33408
outside
1 - 166
Population Genetic Test Statistics
Pi
328.710382
Theta
157.233654
Tajima's D
3.071581
CLR
1.737433
CSRT
0.981900904954752
Interpretation
Uncertain
