Gene
KWMTBOMO14401
Pre Gene Modal
BGIBMGA000112
Annotation
PREDICTED:_cysteine-rich_PDZ-binding_protein_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.11 Extracellular Reliability : 1.135 Nuclear Reliability : 1.624
Sequence
CDS
ATGGTTTGTGAGAAGTGCGAAAAGAAATTGGGCAGAGTAATAACGCCGGACCCTTGGAAGGCTGGAGCTAGGAATACCGTGGAGTCCGGCGGACGTGTTGTTGGAGAAAACAAAGCATTAACCGCCAAAAAAGGACGATTTAATCCGTACACTTCCACATTTCAACAGTGCAAAATCTGCAGAACGAAAGTACACCAGGTCGGATCACATTACTGCCAAGCTTGTGCATACAAAAAGGGTATATGTGCCATGTGTGGAAAGAAGATTCTAGACACTAAGAATTACAGACAGAGCTCAACTTAG
Protein
MVCEKCEKKLGRVITPDPWKAGARNTVESGGRVVGENKALTAKKGRFNPYTSTFQQCKICRTKVHQVGSHYCQACAYKKGICAMCGKKILDTKNYRQSST
Summary
Uniprot
H9IS86
A0A3S2LVM2
A0A1E1W9V4
S4PPF1
A0A212FHS4
A0A194RNN3
+ More
A0A194QH38 D6WGS0 A0A2R7WDI3 A0A310S460 A0A0L7QW81 A0A1B6MS06 A0A1B6GZC1 A0A1B6JAT8 A0A0M9ADR1 A0A2A3E1A9 A0A087ZS03 A0A154P6V7 A0A0T6AY19 A0A0A9XC14 A0A2J7REA8 A0A224XQU5 A0A0V0GBE6 A0A069DNQ4 A0A0J7NUU3 A0A1S3DCH6 A0A2P8ZJN6 A0A232F5P1 K7IYA2 A0A151X3T4 A0A195CZU2 E9IGP0 A0A026W9B3 A0A2C9LMJ6 F4WQT3 A0A195ESP5 A0A158NRS5 E2BHQ2 A0A1B6CXG0 A0A1D2NN15 A0A131Z815 A0A0P4VLW9 R4FLU0 G3MN62 A0A0B6Y717 A0A224Z3X1 A0A1E1XU94 A0A0C9S027 A0A023FEV2 A0A023FW37 A0A0K8RKY8 B7QN05 E2AX55 J3JXN1 R7VLU5 A0A2L2YKM5 K7IXX2 A0A1Y1M663 A0A0L8GZK2 A0A182PD97 A0A182V5L3 A0A182JX94 A0A182WXW0 A0A182I5Y1 A0A182MAA9 A0A1B0GJ20 A0A182WE46 Q7QE02 A0A1S3HZB9 A0A182Y9M6 A0A182RW74 E9GRR6 A0A023EE87 Q16JS4 A0A182FVG5 A0A2M4CYF1 A0A2M4CY91 A0A3B5PRK2 A0A3B5LAL0 A0A084WNL1 A0A182IVF0 A0A3B3YGJ0 A0A087XVV8 A0A3B3V3B2 A0A0S7HY48 A0A1L8E509 A0A3Q1EBE6 A0A3B5B421 A0A3Q1CR79 A0A3P8U374 A0A2P2I5S2 A0A3P9ND08 A0A146Y7H0 A0A1A8PMB1 A0A1A8LV58 A0A1A8JRD3 A0A1A8BBY0 A0A1A8V7U1 A0A146MK50 A0A3R7MBR7
A0A194QH38 D6WGS0 A0A2R7WDI3 A0A310S460 A0A0L7QW81 A0A1B6MS06 A0A1B6GZC1 A0A1B6JAT8 A0A0M9ADR1 A0A2A3E1A9 A0A087ZS03 A0A154P6V7 A0A0T6AY19 A0A0A9XC14 A0A2J7REA8 A0A224XQU5 A0A0V0GBE6 A0A069DNQ4 A0A0J7NUU3 A0A1S3DCH6 A0A2P8ZJN6 A0A232F5P1 K7IYA2 A0A151X3T4 A0A195CZU2 E9IGP0 A0A026W9B3 A0A2C9LMJ6 F4WQT3 A0A195ESP5 A0A158NRS5 E2BHQ2 A0A1B6CXG0 A0A1D2NN15 A0A131Z815 A0A0P4VLW9 R4FLU0 G3MN62 A0A0B6Y717 A0A224Z3X1 A0A1E1XU94 A0A0C9S027 A0A023FEV2 A0A023FW37 A0A0K8RKY8 B7QN05 E2AX55 J3JXN1 R7VLU5 A0A2L2YKM5 K7IXX2 A0A1Y1M663 A0A0L8GZK2 A0A182PD97 A0A182V5L3 A0A182JX94 A0A182WXW0 A0A182I5Y1 A0A182MAA9 A0A1B0GJ20 A0A182WE46 Q7QE02 A0A1S3HZB9 A0A182Y9M6 A0A182RW74 E9GRR6 A0A023EE87 Q16JS4 A0A182FVG5 A0A2M4CYF1 A0A2M4CY91 A0A3B5PRK2 A0A3B5LAL0 A0A084WNL1 A0A182IVF0 A0A3B3YGJ0 A0A087XVV8 A0A3B3V3B2 A0A0S7HY48 A0A1L8E509 A0A3Q1EBE6 A0A3B5B421 A0A3Q1CR79 A0A3P8U374 A0A2P2I5S2 A0A3P9ND08 A0A146Y7H0 A0A1A8PMB1 A0A1A8LV58 A0A1A8JRD3 A0A1A8BBY0 A0A1A8V7U1 A0A146MK50 A0A3R7MBR7
Pubmed
19121390
23622113
22118469
26354079
18362917
19820115
+ More
25401762 26823975 26334808 29403074 28648823 20075255 21282665 24508170 30249741 15562597 21719571 21347285 20798317 27289101 26830274 27129103 22216098 28797301 29209593 26131772 22516182 23537049 23254933 26561354 28004739 12364791 14747013 17210077 25244985 21292972 24945155 17510324 23542700 24438588
25401762 26823975 26334808 29403074 28648823 20075255 21282665 24508170 30249741 15562597 21719571 21347285 20798317 27289101 26830274 27129103 22216098 28797301 29209593 26131772 22516182 23537049 23254933 26561354 28004739 12364791 14747013 17210077 25244985 21292972 24945155 17510324 23542700 24438588
EMBL
BABH01024086
RSAL01000179
RVE44964.1
GDQN01007266
GDQN01005854
JAT83788.1
+ More
JAT85200.1 GAIX01000287 JAA92273.1 AGBW02008471 OWR53267.1 KQ460124 KPJ17626.1 KQ458859 KPJ04858.1 KQ971327 EEZ99622.1 KK854658 PTY17696.1 KQ784160 OAD51865.1 KQ414716 KOC62840.1 GEBQ01001279 JAT38698.1 GECZ01001998 JAS67771.1 GECU01011376 JAS96330.1 KQ435692 KOX81059.1 KZ288453 PBC25533.1 KQ434827 KZC07581.1 LJIG01022593 KRT79749.1 GBHO01027236 GBRD01017066 GDHC01010645 GDHC01004678 JAG16368.1 JAG48761.1 JAQ07984.1 JAQ13951.1 NEVH01005277 PNF39165.1 GFTR01001550 JAW14876.1 GECL01000805 JAP05319.1 GBGD01003598 JAC85291.1 LBMM01001515 KMQ96160.1 PYGN01000035 PSN56714.1 NNAY01000875 OXU26111.1 KQ982562 KYQ54924.1 KQ977041 KYN06127.1 GL763054 EFZ20229.1 KK107347 QOIP01000007 EZA52261.1 RLU20875.1 GL888275 EGI63446.1 KQ981993 KYN30904.1 ADTU01024289 GL448322 EFN84800.1 GEDC01019138 JAS18160.1 LJIJ01000001 ODN06673.1 GEDV01001459 JAP87098.1 GDKW01001109 JAI55486.1 ACPB03004735 GAHY01001253 JAA76257.1 JO843313 AEO34930.1 HACG01005267 CEK52132.1 GFPF01009788 MAA20934.1 GFAA01000592 JAU02843.1 GBZX01002831 JAG89909.1 GBBK01004430 JAC20052.1 GBBL01002390 JAC24930.1 GADI01002609 JAA71199.1 ABJB010114878 DS974685 EEC20227.1 GL443520 EFN62005.1 APGK01042609 BT128001 KB741007 KB632182 AEE62963.1 ENN75611.1 ERL89618.1 AMQN01003920 KB292234 ELU17920.1 IAAA01029724 LAA08663.1 GEZM01039532 JAV81339.1 KQ419780 KOF82393.1 APCN01003537 AXCM01001811 AJWK01019214 AAAB01008848 EAA07057.4 GL732560 EFX77867.1 GAPW01006457 JAC07141.1 CH477994 EAT34537.1 GGFL01006121 MBW70299.1 GGFL01006122 MBW70300.1 ATLV01024620 KE525353 KFB51805.1 AYCK01009265 GBYX01435480 GBYX01435479 JAO45855.1 GFDF01000273 JAV13811.1 IACF01003762 LAB69372.1 GCES01036245 JAR50078.1 HAEH01006816 HAEI01002916 SBR82396.1 HAEF01009288 HAEG01004816 SBR48211.1 HAED01016804 HAEE01002643 SBR22663.1 HADZ01001126 HAEA01016526 SBP65067.1 HADY01003527 HAEJ01015998 SBS56455.1 GCES01166241 JAQ20081.1 QCYY01001511 ROT77561.1
JAT85200.1 GAIX01000287 JAA92273.1 AGBW02008471 OWR53267.1 KQ460124 KPJ17626.1 KQ458859 KPJ04858.1 KQ971327 EEZ99622.1 KK854658 PTY17696.1 KQ784160 OAD51865.1 KQ414716 KOC62840.1 GEBQ01001279 JAT38698.1 GECZ01001998 JAS67771.1 GECU01011376 JAS96330.1 KQ435692 KOX81059.1 KZ288453 PBC25533.1 KQ434827 KZC07581.1 LJIG01022593 KRT79749.1 GBHO01027236 GBRD01017066 GDHC01010645 GDHC01004678 JAG16368.1 JAG48761.1 JAQ07984.1 JAQ13951.1 NEVH01005277 PNF39165.1 GFTR01001550 JAW14876.1 GECL01000805 JAP05319.1 GBGD01003598 JAC85291.1 LBMM01001515 KMQ96160.1 PYGN01000035 PSN56714.1 NNAY01000875 OXU26111.1 KQ982562 KYQ54924.1 KQ977041 KYN06127.1 GL763054 EFZ20229.1 KK107347 QOIP01000007 EZA52261.1 RLU20875.1 GL888275 EGI63446.1 KQ981993 KYN30904.1 ADTU01024289 GL448322 EFN84800.1 GEDC01019138 JAS18160.1 LJIJ01000001 ODN06673.1 GEDV01001459 JAP87098.1 GDKW01001109 JAI55486.1 ACPB03004735 GAHY01001253 JAA76257.1 JO843313 AEO34930.1 HACG01005267 CEK52132.1 GFPF01009788 MAA20934.1 GFAA01000592 JAU02843.1 GBZX01002831 JAG89909.1 GBBK01004430 JAC20052.1 GBBL01002390 JAC24930.1 GADI01002609 JAA71199.1 ABJB010114878 DS974685 EEC20227.1 GL443520 EFN62005.1 APGK01042609 BT128001 KB741007 KB632182 AEE62963.1 ENN75611.1 ERL89618.1 AMQN01003920 KB292234 ELU17920.1 IAAA01029724 LAA08663.1 GEZM01039532 JAV81339.1 KQ419780 KOF82393.1 APCN01003537 AXCM01001811 AJWK01019214 AAAB01008848 EAA07057.4 GL732560 EFX77867.1 GAPW01006457 JAC07141.1 CH477994 EAT34537.1 GGFL01006121 MBW70299.1 GGFL01006122 MBW70300.1 ATLV01024620 KE525353 KFB51805.1 AYCK01009265 GBYX01435480 GBYX01435479 JAO45855.1 GFDF01000273 JAV13811.1 IACF01003762 LAB69372.1 GCES01036245 JAR50078.1 HAEH01006816 HAEI01002916 SBR82396.1 HAEF01009288 HAEG01004816 SBR48211.1 HAED01016804 HAEE01002643 SBR22663.1 HADZ01001126 HAEA01016526 SBP65067.1 HADY01003527 HAEJ01015998 SBS56455.1 GCES01166241 JAQ20081.1 QCYY01001511 ROT77561.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000007266
+ More
UP000053825 UP000053105 UP000242457 UP000005203 UP000076502 UP000235965 UP000036403 UP000079169 UP000245037 UP000215335 UP000002358 UP000075809 UP000078542 UP000053097 UP000279307 UP000076420 UP000007755 UP000078541 UP000005205 UP000008237 UP000094527 UP000015103 UP000001555 UP000000311 UP000019118 UP000030742 UP000014760 UP000053454 UP000075885 UP000075903 UP000075881 UP000076407 UP000075840 UP000075883 UP000092461 UP000075920 UP000007062 UP000085678 UP000076408 UP000075900 UP000000305 UP000008820 UP000069272 UP000002852 UP000261380 UP000030765 UP000075880 UP000261480 UP000028760 UP000261500 UP000257200 UP000261400 UP000257160 UP000265080 UP000242638 UP000265000 UP000283509
UP000053825 UP000053105 UP000242457 UP000005203 UP000076502 UP000235965 UP000036403 UP000079169 UP000245037 UP000215335 UP000002358 UP000075809 UP000078542 UP000053097 UP000279307 UP000076420 UP000007755 UP000078541 UP000005205 UP000008237 UP000094527 UP000015103 UP000001555 UP000000311 UP000019118 UP000030742 UP000014760 UP000053454 UP000075885 UP000075903 UP000075881 UP000076407 UP000075840 UP000075883 UP000092461 UP000075920 UP000007062 UP000085678 UP000076408 UP000075900 UP000000305 UP000008820 UP000069272 UP000002852 UP000261380 UP000030765 UP000075880 UP000261480 UP000028760 UP000261500 UP000257200 UP000261400 UP000257160 UP000265080 UP000242638 UP000265000 UP000283509
PRIDE
ProteinModelPortal
H9IS86
A0A3S2LVM2
A0A1E1W9V4
S4PPF1
A0A212FHS4
A0A194RNN3
+ More
A0A194QH38 D6WGS0 A0A2R7WDI3 A0A310S460 A0A0L7QW81 A0A1B6MS06 A0A1B6GZC1 A0A1B6JAT8 A0A0M9ADR1 A0A2A3E1A9 A0A087ZS03 A0A154P6V7 A0A0T6AY19 A0A0A9XC14 A0A2J7REA8 A0A224XQU5 A0A0V0GBE6 A0A069DNQ4 A0A0J7NUU3 A0A1S3DCH6 A0A2P8ZJN6 A0A232F5P1 K7IYA2 A0A151X3T4 A0A195CZU2 E9IGP0 A0A026W9B3 A0A2C9LMJ6 F4WQT3 A0A195ESP5 A0A158NRS5 E2BHQ2 A0A1B6CXG0 A0A1D2NN15 A0A131Z815 A0A0P4VLW9 R4FLU0 G3MN62 A0A0B6Y717 A0A224Z3X1 A0A1E1XU94 A0A0C9S027 A0A023FEV2 A0A023FW37 A0A0K8RKY8 B7QN05 E2AX55 J3JXN1 R7VLU5 A0A2L2YKM5 K7IXX2 A0A1Y1M663 A0A0L8GZK2 A0A182PD97 A0A182V5L3 A0A182JX94 A0A182WXW0 A0A182I5Y1 A0A182MAA9 A0A1B0GJ20 A0A182WE46 Q7QE02 A0A1S3HZB9 A0A182Y9M6 A0A182RW74 E9GRR6 A0A023EE87 Q16JS4 A0A182FVG5 A0A2M4CYF1 A0A2M4CY91 A0A3B5PRK2 A0A3B5LAL0 A0A084WNL1 A0A182IVF0 A0A3B3YGJ0 A0A087XVV8 A0A3B3V3B2 A0A0S7HY48 A0A1L8E509 A0A3Q1EBE6 A0A3B5B421 A0A3Q1CR79 A0A3P8U374 A0A2P2I5S2 A0A3P9ND08 A0A146Y7H0 A0A1A8PMB1 A0A1A8LV58 A0A1A8JRD3 A0A1A8BBY0 A0A1A8V7U1 A0A146MK50 A0A3R7MBR7
A0A194QH38 D6WGS0 A0A2R7WDI3 A0A310S460 A0A0L7QW81 A0A1B6MS06 A0A1B6GZC1 A0A1B6JAT8 A0A0M9ADR1 A0A2A3E1A9 A0A087ZS03 A0A154P6V7 A0A0T6AY19 A0A0A9XC14 A0A2J7REA8 A0A224XQU5 A0A0V0GBE6 A0A069DNQ4 A0A0J7NUU3 A0A1S3DCH6 A0A2P8ZJN6 A0A232F5P1 K7IYA2 A0A151X3T4 A0A195CZU2 E9IGP0 A0A026W9B3 A0A2C9LMJ6 F4WQT3 A0A195ESP5 A0A158NRS5 E2BHQ2 A0A1B6CXG0 A0A1D2NN15 A0A131Z815 A0A0P4VLW9 R4FLU0 G3MN62 A0A0B6Y717 A0A224Z3X1 A0A1E1XU94 A0A0C9S027 A0A023FEV2 A0A023FW37 A0A0K8RKY8 B7QN05 E2AX55 J3JXN1 R7VLU5 A0A2L2YKM5 K7IXX2 A0A1Y1M663 A0A0L8GZK2 A0A182PD97 A0A182V5L3 A0A182JX94 A0A182WXW0 A0A182I5Y1 A0A182MAA9 A0A1B0GJ20 A0A182WE46 Q7QE02 A0A1S3HZB9 A0A182Y9M6 A0A182RW74 E9GRR6 A0A023EE87 Q16JS4 A0A182FVG5 A0A2M4CYF1 A0A2M4CY91 A0A3B5PRK2 A0A3B5LAL0 A0A084WNL1 A0A182IVF0 A0A3B3YGJ0 A0A087XVV8 A0A3B3V3B2 A0A0S7HY48 A0A1L8E509 A0A3Q1EBE6 A0A3B5B421 A0A3Q1CR79 A0A3P8U374 A0A2P2I5S2 A0A3P9ND08 A0A146Y7H0 A0A1A8PMB1 A0A1A8LV58 A0A1A8JRD3 A0A1A8BBY0 A0A1A8V7U1 A0A146MK50 A0A3R7MBR7
Ontologies
PANTHER
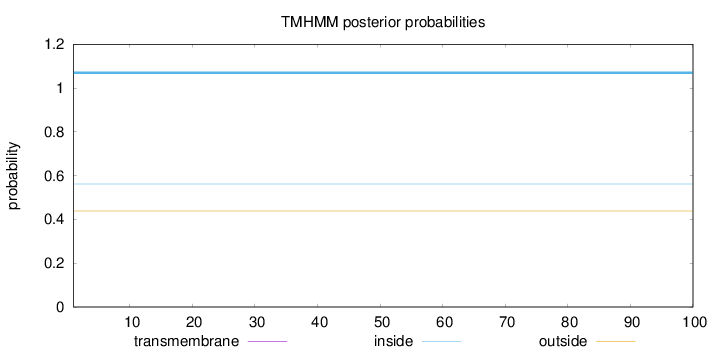
Topology
Length:
100
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00037
Exp number, first 60 AAs:
0
Total prob of N-in:
0.56189
inside
1 - 100
Population Genetic Test Statistics
Pi
21.525277
Theta
24.257457
Tajima's D
-0.429534
CLR
2.291884
CSRT
0.258937053147343
Interpretation
Uncertain
